Abstract
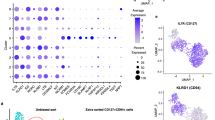
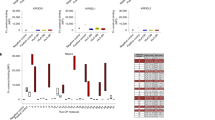
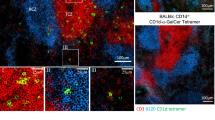
Natural killer (NK) cells are classically viewed as lymphocytes that provide innate surveillance against virally infected cells and tumour cells through the release of cytolytic mediators and interferon (IFN)-γ. In humans, blood CD56dim NK cells specialize in the lysis of cell targets1. In the lymph nodes, CD56bright NK cells secrete IFN-γ cooperating with dendritic cells and T cells in the generation of adaptive responses1,2. Here we report the characterization of a human NK cell subset located in mucosa-associated lymphoid tissues, such as tonsils and Peyer’s patches, which is hard-wired to secrete interleukin (IL)-22, IL-26 and leukaemia inhibitory factor. These NK cells, which we refer to as NK-22 cells, are triggered by acute exposure to IL-23. In vitro, NK-22-secreted cytokines stimulate epithelial cells to secrete IL-10, proliferate and express a variety of mitogenic and anti-apoptotic molecules. NK-22 cells are also found in mouse mucosa-associated lymphoid tissues and appear in the small intestine lamina propria during bacterial infection, suggesting that NK-22 cells provide an innate source of IL-22 that may help constrain inflammation and protect mucosal sites.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cooper, M. A., Fehniger, T. A. & Caligiuri, M. A. The biology of human natural killer-cell subsets. Trends Immunol. 22, 633–640 (2001)
Ferlazzo, G. & Munz, C. NK cell compartments and their activation by dendritic cells. J. Immunol. 172, 1333–1339 (2004)
Vitale, M. et al. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J. Exp. Med. 187, 2065–2072 (1998)
Acosta-Rodriguez, E. V. et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nature Immunol. 8, 639–646 (2007)
Manel, N., Unutmaz, D. & Littman, D. R. The differentiation of human TH-17 cells requires transforming growth factor-β induction of the nuclear receptor RORγt. Nature Immunol. 9, 641–649 (2008)
Candiago, E., Marocolo, D., Manganoni, M. A., Leali, C. & Facchetti, F. Nonlymphoid intraepidermal mononuclear cell collections (pseudo-Pautrier abscesses): a morphologic and immunophenotypical characterization. Am. J. Dermatopathol. 22, 1–6 (2000)
Fuchs, A., Cella, M., Giurisato, E., Shaw, A. S. & Colonna, M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J. Immunol. 172, 3994–3998 (2004)
Cepek, K. L. et al. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the αEβ7 integrin. Nature 372, 190–193 (1994)
Aujla, S. J. et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nature Med. 14, 275–281 (2008)
Zheng, Y. et al. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 445, 648–651 (2007)
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature Med. 14, 282–289 (2008)
Sugimoto, K. et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 118, 534–544 (2008)
Zenewicz, L. A. et al. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity 27, 647–659 (2007)
Hor, S. et al. The T-cell lymphokine interleukin-26 targets epithelial cells through the interleukin-20 receptor 1 and interleukin-10 receptor 2 chains. J. Biol. Chem. 279, 33343–33351 (2004)
Wilson, N. J. et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nature Immunol. 8, 950–957 (2007)
Liang, S. C. et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203, 2271–2279 (2006)
Weaver, C. T., Hatton, R. D., Mangan, P. R. & Harrington, L. E. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25, 821–852 (2007)
Stockinger, B., Veldhoen, M. & Martin, B. Th17 T cells: linking innate and adaptive immunity. Semin. Immunol. 19, 353–361 (2007)
Ouyang, W., Kolls, J. K. & Zheng, Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 28, 454–467 (2008)
Strowig, T. et al. Tonsilar NK cells restrict B cell transformation by the Epstein-Barr virus via IFN-γ. PLoS Pathog. 4, e27 (2008)
Acosta-Rodriguez, E. V., Napolitani, G., Lanzavecchia, A. & Sallusto, F. Interleukins 1β and 6 but not transforming growth factor-β are essential for the differentiation of interleukin 17-producing human T helper cells. Nature Immunol. 8, 942–949 (2007)
Nagalakshmi, M. L., Rascle, A., Zurawski, S., Menon, S. & de Waal Malefyt, R. Interleukin-22 activates STAT3 and induces IL-10 by colon epithelial cells. Int. Immunopharmacol. 4, 679–691 (2004)
Kusam, S. & Dent, A. Common mechanisms for the regulation of B cell differentiation and transformation by the transcriptional repressor protein BCL-6. Immunol. Res. 37, 177–186 (2007)
Veldhoen, M. et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 (2008)
Quintana, F. J. et al. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 (2008)
Brustle, A. et al. The development of inflammatory TH-17 cells requires interferon-regulatory factor 4. Nature Immunol. 8, 958–966 (2007)
Volpe, E. et al. A critical function for transforming growth factor-β, interleukin 23 and proinflammatory cytokines in driving and modulating human TH-17 responses. Nature Immunol. 9, 650–657 (2008)
Zhou, L. et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453, 236–240 (2008)
Yang, L. et al. IL-21 and TGF-β are required for differentiation of human TH17 cells. Nature 454, 350–352 (2008)
Mebius, R. E. Organogenesis of lymphoid tissues. Nature Rev. Immunol. 3, 292–303 (2003)
Acknowledgements
We thank J. Hughes, B. Eades and S. Schloehmann for cell sorting; R. Clary and the nursing staff at the Children’s Hospital, Washington University School of Medicine, for providing tonsil specimens; S. Lonardi for assistance in immunohistochemistry; J. Pfeifer for providing gut specimens; A. Rapaport and S. McCartney for help in gene chip analysis; and S. Gilfillan for critically reading the manuscript. This work was supported by National Institutes of Health (NIH) grants R01AI056139-05 and R21AI067748-02 (to M.Co.) and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant R01 DK079798 (to J.C.M.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH and NIDDK.
Author contributions M.Ce., A.F., K.O. and J.M.D. performed the experiments. W.V., F.F. and J.K.M.L. performed immunohistochemical analyses. J.C.M. designed experiments. M.Co. wrote the manuscript and directed the research.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-11 with Legends, Supplementary Methods and Supplementary Tables 1 and 2 (PDF 1247 kb)
Rights and permissions
About this article
Cite this article
Cella, M., Fuchs, A., Vermi, W. et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009). https://doi.org/10.1038/nature07537
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07537
This article is cited by
-
The emerging family of RORγt+ antigen-presenting cells
Nature Reviews Immunology (2024)
-
Role of IL-22 in intestinal microenvironment and potential targeted therapy through diet
Immunologic Research (2023)
-
Innate lymphoid cells and cancer
Nature Immunology (2022)
-
Interferon regulatory factor 1 (IRF-1) promotes intestinal group 3 innate lymphoid responses during Citrobacter rodentium infection
Nature Communications (2022)
-
Microbiota-derived tryptophan metabolites in vascular inflammation and cardiovascular disease
Amino Acids (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.