Abstract
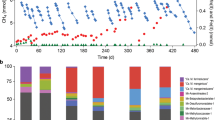
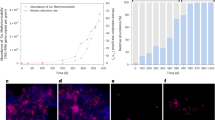
Aerobic methanotrophic bacteria consume methane as it diffuses away from methanogenic zones of soil and sediment1. They act as a biofilter to reduce methane emissions to the atmosphere, and they are therefore targets in strategies to combat global climate change. No cultured methanotroph grows optimally below pH 5, but some environments with active methane cycles are very acidic2,3. Here we describe an extremely acidophilic methanotroph that grows optimally at pH 2.0–2.5. Unlike the known methanotrophs, it does not belong to the phylum Proteobacteria but rather to the Verrucomicrobia, a widespread and diverse bacterial phylum that primarily comprises uncultivated species with unknown genotypes. Analysis of its draft genome detected genes encoding particulate methane monooxygenase that were homologous to genes found in methanotrophic proteobacteria. However, known genetic modules for methanol and formaldehyde oxidation were incomplete or missing, suggesting that the bacterium uses some novel methylotrophic pathways. Phylogenetic analysis of its three pmoA genes (encoding a subunit of particulate methane monooxygenase) placed them into a distinct cluster from proteobacterial homologues. This indicates an ancient divergence of Verrucomicrobia and Proteobacteria methanotrophs rather than a recent horizontal gene transfer of methanotrophic ability. The findings show that methanotrophy in the Bacteria is more taxonomically, ecologically and genetically diverse than previously thought, and that previous studies have failed to assess the full diversity of methanotrophs in acidic environments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Conrad, R. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60, 609–640 (1996)
Castaldi, S. & Tedesco, D. Methane production and consumption in an active volcanic environment of southern Italy. Chemosphere 58, 131–139 (2005)
Dedysh, S. N. et al. Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science 282, 281–284 (1998)
Intergovernmental Panel on Climate Change. Climate Change 2007: The Physical Science Basis. Working Group I Contribution to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change (eds Solomon S. et al.) (Cambridge Univ. Press, Cambridge, 2007)
Kvenvolden, K. A. & Rogers, B. W. Gaia’s breath—global methane exhalations. Mar. Petrol. Geol. 22, 579–590 (2005)
Giggenbach, W. F., Sheppard, D. S., Robinson, B. W., Stewart, M. K. & Lyon, G. L. Geochemical structure and position of the Waiotapu geothermal field, New Zealand. Geothermics 23, 599–644 (1994)
Dedysh, S. N. Methanotrophic bacteria of acidic Sphagnum peat bogs. Microbiology (Russia) 71, 638–650 (2002)
Theisen, A. R. et al. Regulation of methane oxidation in the facultative methanotroph Methylocella silvestris BL2. Mol. Microbiol. 58, 682–692 (2005)
Dumont, M. G. & Murrell, J. C. Community-level analysis: key genes of aerobic methane oxidation. Methods Enzymol. 397, 413–427 (2005)
Knief, C., Lipski, A. & Dunfield, P. F. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 69, 6703–6714 (2003)
Hou, S. et al. Genome sequence of the deep-sea γ-proteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy. Proc. Natl Acad. Sci. USA 101, 18036–18041 (2004)
Ward, N. et al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2, E303 (2004)
Gilbert, B. et al. Molecular analysis of the pmo (particulate methane monooxygenase) operons from two type II methanotrophs. Appl. Environ. Microbiol. 66, 966–975 (2000)
Ricke, P., Erkel, C., Kube, M., Reinhardt, R. & Liesack, W. Comparative analysis of the conventional and novel pmo (particulate methane monooxygenase) operons from Methylocystis strain SC2. Appl. Environ. Microbiol. 70, 3055–3063 (2004)
Holmes, A. J. et al. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 65, 3312–3318 (1999)
Chistoserdova, L., Chen, S. W., Lapidus, A. & Lidstrom, M. L. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185, 2980–2987 (2003)
Chistoserdova, L. et al. Genome of Methylobacillus flagellatus, molecular basis for obligate methylotrophy, and polyphyletic origin of methylotrophy. J. Bacteriol. 189, 4020–4027 (2007)
Kane, S. R. et al. Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibium petroleiphilum PM1. J. Bacteriol. 189, 1931–1945 (2007)
Chistoserdova, L. et al. The enigmatic Planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol. Biol. Evol. 21, 1234–1241 (2004)
Trotsenko, Y. Metabolic features of methane- and methanol-utilizing bacteria. Acta Biotechnol. 3, 269–277 (2004)
Kelly, D. P., Anthony, C. & Murrell, J. C. Insights from the complete genome sequence of the obligate methanotroph, Methylococcus capsulatus . Trends Microbiol. 13, 195–198 (2005)
Janssen, P. H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728 (2006)
Bryant, D. A. et al. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic Acidobacterium . Science 317, 523–526 (2007)
Morris, S. A., Radajewski, S., Willison, T. W. & Murrell, J. C. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68, 1446–1453 (2002)
Lin, J. L. et al. Molecular diversity of methanotrophs in Transbaikal soda lake sediments and identification of potentially active populations by stable isotope probing. Environ. Microbiol. 6, 1049–1060 (2004)
Dedysh, S. N. et al. Methylocystis heyerii sp. nov., a novel type II methanotrophic bacterium possessing the ‘signature’ fatty acid of type I methanotrophs. Int. J. Syst. Evol. Microbiol. 57, 472–479 (2007)
Weatherby, T. M. & Lenz, P. H. Mechanoreceptors in calanoid copepods: Designed for high sensitivity. Arthropod Struct. Dev. 29, 275–288 (2000)
Ispolatov, I., Yuryev, A., Mazo, I. & Maslov, S. Binding properties and evolution of homodimers in protein–protein interaction networks. Nucleic Acids Res. 33, 3629–3635 (2005)
Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002)
Acknowledgements
We thank S. Dedysh, J. C. Murrell and J. Euzéby for comments; A. Malahoff for supporting this research programme; and the Tikitere Trust for permission to sample at Hell’s Gate. This work was supported in part by the Wairakei Environmental Mitigation Charitable Trust (P.D.). The genome analysis was funded by the US Department of Defense (M.A.). Gene sequences referenced in this paper are deposited DDBJ/EMBL/GenBank under accession numbers AM900833–AM900834 and EU223838–EU223931.
Author Contributions P.F.D., B.W.M., M.A.C. and M.B.S. performed field sampling, methane measurement and molecular 16S rRNA analyses. P.F.D. and M.B.S. isolated and characterized the culture. S.H., B.L., J.H.S., Z.Z., Y.R., J.W., L.F., M.B.S., L.W., W.L. and M.A. conducted genome sequencing. P.F.D., P.S., A.Y., A.V.S., J.S., P.S. and M.A. conducted genome analyses. T.M.W. and M.B.S. performed electron microscopy. P.B. undertook phospholipid fatty-acid analysis.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figure 1 with Legend and Supplementary Tables 1-3. The Supplementary Figure 1 shows phylogenetic 16S rRNA gene tree showing the position of isolate V4 relative to other members of the phylum Verrucomicrobia and to proteobacterial methanotrophs. The Supplementary Table 1 illustrates putative genes involved in one-carbon metabolism in Verrucomicrobia isolate V4. The Supplementary Table 2 illustrates identities of partial segments (165 amino acids) of derived PmoA sequences in isolate V4 to PmoA and AmoA sequences of selected bacterial nitrifiers and methanotrophs, and the occurrence of proposed "signature" amino acid residues for either PmoA or AmoA. The Supplementary Table 3 illustrates phospholipid fatty acid profile of isolate V4. (PDF 479 kb)
Rights and permissions
About this article
Cite this article
Dunfield, P., Yuryev, A., Senin, P. et al. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450, 879–882 (2007). https://doi.org/10.1038/nature06411
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06411
This article is cited by
-
Simultaneous sulfide and methane oxidation by an extremophile
Nature Communications (2023)
-
Microbial Diversity of Deep-sea Sediments from Three Newly Discovered Hydrothermal Vent Fields in the Central Indian Ridge
Ocean Science Journal (2023)
-
Universal activity-based labeling method for ammonia- and alkane-oxidizing bacteria
The ISME Journal (2022)
-
Prospecting the significance of methane-utilizing bacteria in agriculture
World Journal of Microbiology and Biotechnology (2022)
-
Methane oxidation in a landfill biowindow under wide seasonally fluctuating climatic conditions
Environmental Science and Pollution Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.