Abstract
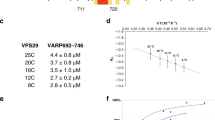
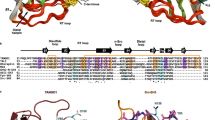
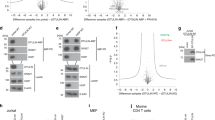
The retromer complex1,2 is required for the sorting of acid hydrolases to lysosomes3,4,5,6,7, transcytosis of the polymeric immunoglobulin receptor8, Wnt gradient formation9,10, iron transporter recycling11 and processing of the amyloid precursor protein12. Human retromer consists of two smaller complexes: the cargo recognition VPS26–VPS29–VPS35 heterotrimer and a membrane-targeting heterodimer or homodimer of SNX1 and/or SNX2 (ref. 13). Here we report the crystal structure of a VPS29–VPS35 subcomplex showing how the metallophosphoesterase-fold subunit VPS29 (refs 14, 15) acts as a scaffold for the carboxy-terminal half of VPS35. VPS35 forms a horseshoe-shaped, right-handed, α-helical solenoid, the concave face of which completely covers the metal-binding site of VPS29, whereas the convex face exposes a series of hydrophobic interhelical grooves. Electron microscopy shows that the intact VPS26–VPS29–VPS35 complex is a stick-shaped, flexible structure, approximately 21 nm long. A hybrid structural model derived from crystal structures, electron microscopy, interaction studies and bioinformatics shows that the α-solenoid fold extends the full length of VPS35, and that VPS26 is bound at the opposite end from VPS29. This extended structure presents multiple binding sites for the SNX complex and receptor cargo, and appears capable of flexing to conform to curved vesicular membranes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Seaman, M. N. Recycle your receptors with retromer. Trends Cell Biol. 15, 68–75 (2005)
Bonifacino, J. S. & Rojas, R. Retrograde transport from endosomes to the trans-Golgi network. Nature Rev. Mol. Cell Biol. 7, 568–579 (2006)
Seaman, M. N. J., McCaffery, J. M. & Emr, S. D. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142, 665–681 (1998)
Haft, C. R. et al. Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: Assembly into multimeric complexes. Mol. Biol. Cell 11, 4105–4116 (2000)
Arighi, C. N., Hartnell, L. M., Aguilar, R. C., Haft, C. R. & Bonifacino, J. S. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165, 123–133 (2004)
Carlton, J. et al. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr. Biol. 14, 1791–1800 (2004)
Seaman, M. N. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165, 111–122 (2004)
Verges, M. et al. The mammalian retromer regulates transcytosis of the polymeric immunoglobulin receptor. Nature Cell Biol. 6, 763–769 (2004)
Coudreuse, D. Y. M., Roel, G., Betist, M. C., Destree, O. & Korswagen, H. C. Wnt gradient formation requires retromer function in Wnt-producing cells. Science 312, 921–924 (2006)
Prasad, B. C. & Clark, S. G. Wnt signaling establishes anteroposterior neuronal polarity and requires retromer in C. elegans. . Development 133, 1757–1766 (2006)
Strochlic, T. I., Setty, T. G., Sitaram, A. & Burd, C. G. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 177, 115–125 (2007)
Small, S. A. et al. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann. Neurol. 58, 909–919 (2005)
Rojas, R., Kametaka, S., Haft, C. R. & Bonifacino, J. S. Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of the mannose 6-phosphate receptors. Mol. Cell. Biol. 27, 1112–1124 (2007)
Wang, D. et al. Crystal structure of human vacuolar protein sorting protein 29 reveals a phosphodiesterase/nuclease-like fold and two protein-protein interaction sites. J. Biol. Chem. 280, 22962–22967 (2005)
Collins, B. M., Skinner, C. F., Watson, P. J., Seaman, M. N. & Owen, D. J. Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nature Struct. Mol. Biol. 12, 594–602 (2005)
Shi, H., Rojas, R., Bonifacino, J. S. & Hurley, J. H. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nature Struct. Mol. Biol. 13, 540–548 (2006)
Damen, E., Krieger, E., Nielsen, J. E., Eygensteyn, J. & van Leeuwen, J. E. M. The human Vps29 retromer component is a metallo-phosphoesterase for a cation-independent mannose 6-phosphate receptor substrate peptide. Biochem. J. 398, 399–409 (2006)
Collins, B. M., McCoy, A. J., Kent, H. M., Evans, P. R. & Owen, D. J. Molecular architecture and functional model of the endocytic AP2 complex. Cell 109, 523–535 (2002)
Heldwein, E. E. et al. Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl Acad. Sci. USA 101, 14108–14113 (2004)
Ybe, J. A. et al. Clathrin self-assembly is mediated by a tandemly repeated superhelix. Nature 399, 371–375 (1999)
Fotin, A. et al. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature 432, 573–579 (2004)
Fath, S., Mancias, J. D., Bi, X. & Goldberg, J. Structure and organization of coat proteins in the COPII cage. Cell 129, 1251–1252 (2007)
Misra, S., Puertollano, R., Kato, Y., Bonifacino, J. S. & Hurley, J. H. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415, 933–937 (2002)
Shiba, T. et al. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature 415, 937–941 (2002)
Nothwehr, S. F., Bruinsma, P. & Strawn, L. A. Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol. Biol. Cell 10, 875–890 (1999)
Yang, J. et al. Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J. 24, 1–10 (2005)
Groves, M. R. & Barford, D. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9, 383–389 (1999)
Conti, E. & Izaurralde, E. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 13, 310–319 (2001)
Nothwehr, S. F., Ha, S. A. & Bruinsma, P. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J. Cell Biol. 151, 297–310 (2000)
Peter, B. J. et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 303, 495–499 (2004)
Sheffield, P., Garrard, S. & Derewenda, Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15, 34–39 (1999)
Hierro, A., Kim, J. & Hurley, J. H. Polycistronic expression and purification of the ESCRT-II endosomal trafficking complex. Methods Enzymol. 403, 322–332 (2005)
Tan, S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli . Protein Expr. Purif. 21, 224–234 (2001)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Pettit, F. K., Bare, E., Tsai, A. & Bowie, J. U. HotPatch: a statistical approach to finding biologically relevant features on protein surfaces. J. Mol. Biol. 369, 863–879 (2007)
Bateman, A. et al. The Pfam protein families database. Nucleic Acids Res. 30, 276–280 (2002)
Martina, J. A., Moriyama, K. & Bonifacino, J. S. BLOC-3, a protein complex containing the Hermansky-Pudlak syndrome gene products HPS1 and HPS4. J. Biol. Chem. 278, 29376–29384 (2003)
Heymann, J. B. & Belnap, D. M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007)
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Bucher, P., Karplus, K., Moeri, N. & Hofmann, K. A flexible motif search technique based on generalized profiles. Comput. Chem. 20, 3–23 (1996)
Hofmann, K. & Bucher, P. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem. Sci. 20, 347–349 (1995)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Acknowledgements
We thank H. Shi for contributing to the early stages of this project; X. Zhu and H. T. Tsai for technical assistance; B. Canagarajah for bioinformatics and computational crystallography support; E. Tyler for artwork; Z.-Q. Fu and all the staff of APS SER-CAT beamline 22 for assistance with data collection; and C. Haft for antibodies to retromer. Use of the APS was supported by the US DOE, Basic Energy Sciences, Office of Science. This project was funded by the Intramural Programs of NIDDK, NICHD and NIAMS, NIH.
Author Contributions A.L.R. and A.H. expressed and purified protein complexes, crystallized the VPS29–VPS35 C-terminal subcomplex, collected crystallographic data, and determined and refined the crystal structure; A.H. carried out phosphatase assays; R.R. and N.M. carried out immunoprecipitation and optical microscopy studies; G.E. and A.C.S. carried out and interpreted electron microscopy studies; A.V.K. carried out sequence analysis; and J.H.H., J.S.B. and A.C.S. designed the study. A.H. and A.L.R. contributed equally to this study.
Crystallographic coordinates have been deposited with the Protein Data Bank with accession number 2R17.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Crystallographic coordinates have been deposited with the Protein Data Bank with accession number 2R17. Reprints and permissions information is available at www.nature.com/reprints.
Supplementary information
Supplementary Information
The file contains Supplementary Figures S1-S8 with Legends and Supplementary Tables S1-S2. (PDF 5515 kb)
Rights and permissions
About this article
Cite this article
Hierro, A., Rojas, A., Rojas, R. et al. Functional architecture of the retromer cargo-recognition complex. Nature 449, 1063–1067 (2007). https://doi.org/10.1038/nature06216
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06216
This article is cited by
-
VPS35 promotes gastric cancer progression through integrin/FAK/SRC signalling-mediated IL-6/STAT3 pathway activation in a YAP-dependent manner
Oncogene (2024)
-
Nanobody-based VSR7 tracing shows clathrin-dependent TGN to Golgi recycling
Nature Communications (2023)
-
Retromer subunit, CfVps35 is required for growth development and pathogenicity of Colletotrichum fructicola
BMC Genomic Data (2022)
-
ITRAQ-based quantitative proteomic analysis reveals that VPS35 promotes the expression of MCM2-7 genes in HeLa cells
Scientific Reports (2022)
-
Identification of specific role of SNX family in gastric cancer prognosis evaluation
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.