Abstract
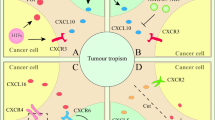
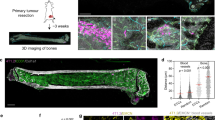
Mesenchymal stem cells have been recently described to localize to breast carcinomas, where they integrate into the tumour-associated stroma. However, the involvement of mesenchymal stem cells (or their derivatives) in tumour pathophysiology has not been addressed. Here, we demonstrate that bone-marrow-derived human mesenchymal stem cells, when mixed with otherwise weakly metastatic human breast carcinoma cells, cause the cancer cells to increase their metastatic potency greatly when this cell mixture is introduced into a subcutaneous site and allowed to form a tumour xenograft. The breast cancer cells stimulate de novo secretion of the chemokine CCL5 (also called RANTES) from mesenchymal stem cells, which then acts in a paracrine fashion on the cancer cells to enhance their motility, invasion and metastasis. This enhanced metastatic ability is reversible and is dependent on CCL5 signalling through the chemokine receptor CCR5. Collectively, these data demonstrate that the tumour microenvironment facilitates metastatic spread by eliciting reversible changes in the phenotype of cancer cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bissell, M. J. & Radisky, D. Putting tumours in context. Nature Rev. Cancer 1, 46–54 (2001)
Hall, B., Andreeff, M. & Marini, F. The participation of mesenchymal stem cells in tumor stroma formation and their application as targeted-gene delivery vehicles. Handb. Exp. Pharmacol. 180, 263–283 (2007)
Pittenger, M. F. et al. Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147 (1999)
Young, H. E. et al. Clonogenic analysis reveals reserve stem cells in postnatal mammals: I. Pluripotent mesenchymal stem cells. Anat. Rec. 263, 350–360 (2001)
Young, H. E. et al. Human reserve pluripotent mesenchymal stem cells are present in the connective tissues of skeletal muscle and dermis derived from fetal, adult, and geriatric donors. Anat. Rec. 264, 51–62 (2001)
Fox, J. M., Chamberlain, G., Ashton, B. A. & Middleton, J. Recent advances into the understanding of mesenchymal stem cell trafficking. Br. J. Haematol. 137, 491–502 (2007)
Gregory, C. A., Prockop, D. J. & Spees, J. L. Non-hematopoietic bone marrow stem cells: molecular control of expansion and differentiation. Exp. Cell Res. 306, 330–335 (2005)
Park, C. C., Bissell, M. J. & Barcellos-Hoff, M. H. The influence of the microenvironment on the malignant phenotype. Mol. Med. Today 6, 324–329 (2000)
Nakamura, K. et al. Antitumor effect of genetically engineered mesenchymal stem cells in a rat glioma model. Gene Ther. 11, 1155–1164 (2004)
Nakamizo, A. et al. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res. 65, 3307–3318 (2005)
Hung, S. C. et al. Mesenchymal stem cell targeting of microscopic tumors and tumor stroma development monitored by noninvasive in vivo positron emission tomography imaging. Clin. Cancer Res. 11, 7749–7756 (2005)
Menon, L. G. et al. Differential gene expression associated with migration of mesenchymal stem cells to conditioned medium from tumor cells or bone marrow cells. Stem Cells 25, 520–528 (2007)
Komarova, S., Kawakami, Y., Stoff-Khalili, M. A., Curiel, D. T. & Pereboeva, L. Mesenchymal progenitor cells as cellular vehicles for delivery of oncolytic adenoviruses. Mol. Cancer Ther. 5, 755–766 (2006)
Khakoo, A. Y. et al. Human mesenchymal stem cells exert potent antitumorigenic effects in a model of Kaposi’s sarcoma. J. Exp. Med. 203, 1235–1247 (2006)
Studeny, M. et al. Bone marrow-derived mesenchymal stem cells as vehicles for interferon-β delivery into tumors. Cancer Res. 62, 3603–3608 (2002)
Luboshits, G. et al. Elevated expression of the CC chemokine regulated on activation, normal T cell expressed and secreted (RANTES) in advanced breast carcinoma. Cancer Res. 59, 4681–4687 (1999)
Niwa, Y. et al. Correlation of tissue and plasma RANTES levels with disease course in patients with breast or cervical cancer. Clin. Cancer Res. 7, 285–289 (2001)
Azenshtein, E. et al. The CC chemokine RANTES in breast carcinoma progression: regulation of expression and potential mechanisms of promalignant activity. Cancer Res. 62, 1093–1102 (2002)
Robinson, S. C. et al. A chemokine receptor antagonist inhibits experimental breast tumor growth. Cancer Res. 63, 8360–8365 (2003)
Hillyer, P. & Male, D. Expression of chemokines on the surface of different human endothelia. Immunol. Cell Biol. 83, 375–382 (2005)
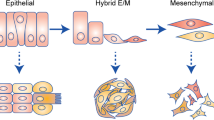
Thiery, J. P. Epithelial-mesenchymal transitions in tumour progression. Nature Rev. Cancer 2, 442–454 (2002)
Toker, A. & Yoeli-Lerner, M. Akt signaling and cancer: surviving but not moving on. Cancer Res. 66, 3963–3966 (2006)
Tanaka, T. et al. Chemokines in tumor progression and metastasis. Cancer Sci. 96, 317–322 (2005)
Mira, E. et al. A role for chemokine receptor transactivation in growth factor signaling. EMBO Rep. 2, 151–156 (2001)
Qin, X. F., An, D. S., Chen, I. S. & Baltimore, D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl Acad. Sci. USA 100, 183–188 (2003)
West, R. B. et al. Determination of stromal signatures in breast carcinoma. PLoS Biol. 3, e187 (2005)
Allinen, M. et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 6, 17–32 (2004)
Balkwill, F. Cancer and the chemokine network. Nature Rev. Cancer 4, 540–550 (2004)
Karnoub, A. E. & Weinberg, R. A. Chemokine networks and breast cancer metastasis. Breast Dis. 26, 75–85 (2006)
Palani, A. & Tagat, J. R. Discovery and development of small-molecule chemokine coreceptor CCR5 antagonists. J. Med. Chem. 49, 2851–2857 (2006)
Orimo, A. et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121, 335–348 (2005)
Elenbaas, B. et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 15, 50–65 (2001)
Lodie, T. A. et al. Systematic analysis of reportedly distinct populations of multipotent bone marrow-derived stem cells reveals a lack of distinction. Tissue Eng. 8, 739–751 (2002)
Hahn, W. C. et al. Creation of human tumour cells with defined genetic elements. Nature 400, 464–468 (1999)
Kuperwasser, C. et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc. Natl Acad. Sci. USA 101, 4966–4971 (2004)
Richardson, A. L. et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9, 121–132 (2006)
Acknowledgements
We thank F. Reinhardt for assistance in animal studies, A. Lu for technical help, J. Yao for SAGE data analysis and the MIT Comparative Pathology Laboratory for immunohistochemical analyses. We are grateful to A. Bernad, X.-F. Qin, D. Baltimore and W. Hahn for providing constructs. We would also like to thank R. Hynes, T. Jacks and R. Goldsby for discussions. A.E.K. is a fellow of the Susan G. Komen Breast Cancer Foundation. R.A.W. is an American Cancer Society Research Professor and a Daniel K. Ludwig Cancer Research Professor. This research is supported by grants from the Breast Cancer Research Foundation (R.A.W.), the Ludwig Trust (R.A.W.), the Susan G. Komen Breast Cancer Foundation (R.A.W.) and the Dana-Farber/Harvard Cancer Center Specialized Program of Research Excellence (SPORE) in Breast Cancer (A.E.K., R.A.W. and K.P.).
Author Contributions A.E.K. conceived and designed this study, and performed most experiments; R.A.W. supervised research; A.E.K. and R.A.W. wrote the manuscript; A.B.D. and R.T. provided human MSCs; A.B.D. helped in in vivo CCL5 neutralization; A.S. helped in the Luminex screens; A.P.V. and M.W.B. provided technical support in tissue culture, ELISA, western blot, RT–PCR and soft-agar analyses; G.W.B. performed CCL5 analysis on soft tumour expression data; A.L.R. obtained and classified the clinical specimens; K.P. fractionated the clinical samples and performed SAGE analyses; and A.L.R. performed the microarray analysis on sorted stroma.
The clinical microarray data on the sorted stroma is deposited at http://www.ncbi.nlm.nih.gov/geo, GSE8977
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-9 and Supplementary Table 1. (PDF 1132 kb)
Rights and permissions
About this article
Cite this article
Karnoub, A., Dash, A., Vo, A. et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449, 557–563 (2007). https://doi.org/10.1038/nature06188
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06188
This article is cited by
-
Tumor-derived small extracellular vesicles facilitate omental metastasis of ovarian cancer by triggering activation of mesenchymal stem cells
Cell Communication and Signaling (2024)
-
Evidence of steady-state fibroblast subtypes in the normal human breast as cells-of-origin for perturbed-state fibroblasts in breast cancer
Breast Cancer Research (2024)
-
ZO-1 regulates the migration of mesenchymal stem cells in cooperation with α-catenin in response to breast tumor cells
Cell Death Discovery (2024)
-
Cell fusion upregulates PD-L1 expression for evasion from immunosurveillance
Cancer Gene Therapy (2024)
-
Exosomes released by oxidative stress-induced mesenchymal stem cells promote murine mammary tumor progression through activating the STAT3 signaling pathway
Molecular and Cellular Biochemistry (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.