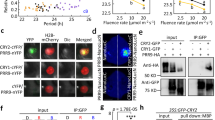
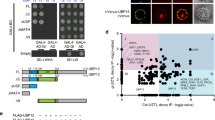
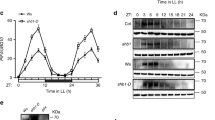
Abstract
The circadian clock is essential for coordinating the proper phasing of many important cellular processes. Robust cycling of key clock elements is required to maintain strong circadian oscillations of these clock-controlled outputs. Rhythmic expression of the Arabidopsis thaliana F-box protein ZEITLUPE (ZTL) is necessary to sustain a normal circadian period by controlling the proteasome-dependent degradation of a central clock protein, TIMING OF CAB EXPRESSION 1 (TOC1)1,2. ZTL messenger RNA is constitutively expressed, but ZTL protein levels oscillate with a threefold change in amplitude through an unknown mechanism3. Here we show that GIGANTEA (GI) is essential to establish and sustain oscillations of ZTL by a direct protein–protein interaction. GI, a large plant-specific protein with a previously undefined molecular role, stabilizes ZTL in vivo. Furthermore, the ZTL–GI interaction is strongly and specifically enhanced by blue light, through the amino-terminal flavin-binding LIGHT, OXYGEN OR VOLTAGE (LOV) domain of ZTL. Mutations within this domain greatly diminish ZTL–GI interactions, leading to strongly reduced ZTL levels. Notably, a C82A mutation in the LOV domain, implicated in the flavin-dependent photochemistry, eliminates blue-light-enhanced binding of GI to ZTL. These data establish ZTL as a blue-light photoreceptor, which facilitates its own stability through a blue-light-enhanced GI interaction. Because the regulation of GI transcription is clock-controlled, consequent GI protein cycling confers a post-translational rhythm on ZTL protein. This mechanism of establishing and sustaining robust oscillations of ZTL results in the high-amplitude TOC1 rhythms necessary for proper clock function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
20 September 2007
In the AOP version of this manuscript the LR and LB panels of ZTL in Fig. 3c were inadvertently inverted; this was corrected on 20 September 2007
References
Somers, D. E., Schultz, T. F., Milnamow, M. & Kay, S. A. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329 (2000)
Mas, P., Kim, W. Y., Somers, D. E. & Kay, S. A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570 (2003)
Kim, W. Y., Geng, R. & Somers, D. E. Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl Acad. Sci. USA 100, 4933–4938 (2003)
Fowler, S. et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688 (1999)
Park, D. et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582 (1999)
Martin-Tryon, E. L., Kreps, J. A. & Harmer, S. L. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143, 473–486 (2007)
Gould, P. D. et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18, 1177–1187 (2006)
David, K. M., Armbruster, U., Tama, N. & Putterill, J. Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett. 580, 1193–1197 (2006)
Nelson, D. C., Lasswell, J., Rogg, L. E., Cohen, M. A. & Bartel, B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340 (2000)
Schultz, T. F., Kiyosue, T., Yanovsky, M., Wada, M. & Kay, S. A. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13, 2659–2670 (2001)
Zeng, L. R. et al. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808 (2004)
Crosson, S. & Moffat, K. Structure of a flavin-binding plant photoreceptor domain: Insights into light-mediated signal transduction. Proc. Natl Acad. Sci. USA 98, 2995–3000 (2001)
Froehlich, A. C., Liu, Y., Loros, J. J. & Dunlap, J. C. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297, 815–819 (2002)
He, Q. et al. White Collar-1, a DNA binding transcription factor and a light sensor. Science 297, 840–843 (2002)
Imaizumi, T., Tran, H. G., Swartz, T. E., Briggs, W. R. & Kay, S. A. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306 (2003)
Voinnet, O., Rivas, S., Mestre, P. & Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956 (2003)
Kevei, E. et al. Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol. 140, 933–945 (2006)
Han, L., Mason, M., Risseeuw, E. P., Crosby, W. L. & Somers, D. E. Formation of an SCF complex is required for proper regulation of circadian timing. Plant J. 40, 291–301 (2004)
Somers, D. E., Kim, W. Y. & Geng, R. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16, 769–782 (2004)
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A. & Kay, S. A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297 (2005)
Salomon, M., Christie, J. M., Knieb, E., Lempert, U. & Briggs, W. R. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39, 9401–9410 (2000)
Strayer, C. et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771 (2000)
Harper, S. M., Neil, L. C. & Gardner, K. H. Structural basis of a phototropin light switch. Science 301, 1541–1544 (2003)
Christie, J. M. et al. Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282, 1698–1701 (1998)
Lee, C., Etchegaray, J. P., Cagampang, F. R., Loudon, A. S. & Reppert, S. M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867 (2001)
Cardone, L. et al. Circadian clock control by SUMOylation of BMAL1. Science 309, 1390–1394 (2005)
Mizoguchi, T. et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270 (2005)
Cheng, X. F. & Wang, Z. Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 43, 758–768 (2005)
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998)
Rubio, V. et al. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41, 767–778 (2005)
Seki, M. et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296, 141–145 (2002)
Seki, M., Carninci, P., Nishiyama, Y., Hayashizaki, Y. & Shinozaki, K. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15, 707–720 (1998)
Ryu, J. S. et al. Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120, 395–406 (2005)
Acknowledgements
We thank S. L. Harmer for providing ztl-103 in advance of publication, S. Kay for TOC1-TOC1-YFP and LKP2-OX seed, D. Bisaro for the adenosine kinase antibody and J. Na for excellent technical assistance. T. Mizoguchi provided gi-3, gi-4, gi-5, gi-6, and GI-OX (Ler). W.-Y. K. was supported in part by a grant from the MOST/KOSEF to the Environmental Biotechnology National Core Research Center (BK21 program grant), Korea. S.F. was funded in part by Support for Long-term Visit from the Yamada Science Foundation. Support is acknowledged from a Korea Science and Engineering Foundation (KOSEF) grant funded by MOST and the National Core Research Center for Systems Biodynamics (to H.G.N.), from the NZ Marsden grant (to J.P.), and from the NSF (IBN and MCB) to D.E.S.
Author Contributions W.-Y.K., S.F., S.-S.S., L.H., J.K. and Y.K. performed experiments. W.-Y.K. and H.-G.N. co-directed and designed experiments. J.P., K.D. and H.G.N. contributed novel reagents. J.P., S.F. and H.G.N. commented on and edited the manuscript. D.E.S. directed research, designed experiments and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures S1-S6 with Legends. (PDF 534 kb)
Rights and permissions
About this article
Cite this article
Kim, WY., Fujiwara, S., Suh, SS. et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360 (2007). https://doi.org/10.1038/nature06132
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06132
This article is cited by
-
Genome-wide identification and characterization of flowering genes in Citrus sinensis (L.) Osbeck: a comparison among C. Medica L., C. Reticulata Blanco, C. Grandis (L.) Osbeck and C. Clementina
BMC Genomic Data (2024)
-
Disrupting FKF1 homodimerization increases FT transcript levels in the evening by enhancing CO stabilization
Plant Cell Reports (2024)
-
Characterizing the circadian connectome of Ocimum tenuiflorum using an integrated network theoretic framework
Scientific Reports (2023)
-
Dim artificial light at night alters gene expression rhythms and growth in a key seagrass species (Posidonia oceanica)
Scientific Reports (2023)
-
Flowering in sugarcane-insights from the grasses
3 Biotech (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.