Abstract
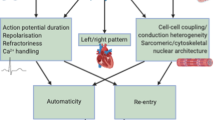
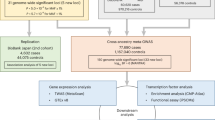
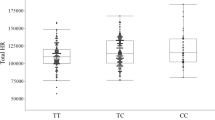
Atrial fibrillation (AF) is the most common sustained cardiac arrhythmia in humans and is characterized by chaotic electrical activity of the atria1. It affects one in ten individuals over the age of 80 years, causes significant morbidity and is an independent predictor of mortality2. Recent studies have provided evidence of a genetic contribution to AF3,4,5. Mutations in potassium-channel genes have been associated with familial AF6,7,8,9,10 but account for only a small fraction of all cases of AF11,12. We have performed a genome-wide association scan, followed by replication studies in three populations of European descent and a Chinese population from Hong Kong and find a strong association between two sequence variants on chromosome 4q25 and AF. Here we show that about 35% of individuals of European descent have at least one of the variants and that the risk of AF increases by 1.72 and 1.39 per copy. The association with the stronger variant is replicated in the Chinese population, where it is carried by 75% of individuals and the risk of AF is increased by 1.42 per copy. A stronger association was observed in individuals with typical atrial flutter. Both variants are adjacent to PITX2, which is known to have a critical function in left–right asymmetry of the heart13,14,15.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Go, A. S. et al. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the AnTicoagulation and Risk Factors in Atrial Fibrillation (ATRIA) Study. J. Am. Med. Assoc. 285, 2370–2375 (2001)
Miyasaka, Y. et al. Secular trends in incidence of atrial fibrillation in Olmsted County, Minnesota, 1980 to 2000, and implications on the projections for future prevalence. Circulation 114, 119–125 (2006)
Arnar, D. O. et al. Familial aggregation of atrial fibrillation in Iceland. Eur. Heart J. 27, 708–712 (2006)
Fox, C. S. et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. J. Am. Med. Assoc. 291, 2851–2855 (2004)
Ellinor, P. T., Yoerger, D. M., Ruskin, J. N. & MacRae, C. A. Familial aggregation in lone atrial fibrillation. Hum. Genet. 118, 179–184 (2005)
Chen, Y. H. et al. KCNQ1 gain-of-function mutation in familial fibrillation. Science 299, 251–254 (2003)
Yang, Y. et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am. J. Hum. Genet. 75, 899–905 (2004)
Xia, M. et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem. Biophys. Res. Commun. 332, 1012–1019 (2005)
Olson, T. M. et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum. Mol. Genet. 15, 2185–2191 (2006)
Hong, K., Bjerregaard, P., Gussak, I. & Brugada, R. Short QT syndrome and atrial fibrillation caused by mutation in KCNH2. J. Cardiovasc. Electrophysiol. 16, 394–396 (2005)
Ellinor, P. T. et al. Mutations in the long QT gene, KCNQ1, are an uncommon cause of atrial fibrillation. Heart 90, 1487–1488 (2004)
Ellinor, P. T., Petrov-Kondratov, V. I., Zakharova, E., Nam, E. G. & MacRae, C. A. Potassium channel gene mutations rarely cause atrial fibrillation. BMC Med. Genet. 7, 70 (2006)
Franco, D. & Campione, M. The role of Pitx2 during cardiac development. Linking left–right signaling and congenital heart diseases. Trends Cardiovasc. Med. 13, 157–163 (2003)
Faucourt, M., Houliston, E., Besnardeau, L., Kimelman, D. & Lepage, T. The pitx2 homeobox protein is required early for endoderm formation and nodal signaling. Dev. Biol. 229, 287–306 (2001)
Mommersteeg, M. T. et al. Molecular pathway for the localized formation of the sinoatrial node. Circ. Res. 100, 354–362 (2007)
The International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005)
Waldo, A. L. The interrelationship between atrial fibrillation and atrial flutter. Prog. Cardiovasc. Dis. 48, 41–56 (2005)
Zini, S. et al. Identification of metabolic pathways of brain angiotensin II and III using specific aminopeptidase inhibitors: predominant role of angiotensin III in the control of vasopressin release. Proc. Natl Acad. Sci. USA 93, 11968–11973 (1996)
Gretarsdottir, S. et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nature Genet. 35, 131–138 (2003)
Falk, C. T. & Rubinstein, P. Haplotype relative risks: an easy reliable way to construct a proper control sample for risk calculations. Ann. Hum. Genet. 51, 227–233 (1987)
Mantel, N. & Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748 (1959)
Grant, S. F. et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genet. 38, 320–323 (2006)
Yang, X. et al. Development and validation of stroke risk equation for Hong Kong Chinese patients with type 2 diabetes: the Hong Kong Diabetes Registry. Diabetes Care 30, 65–70 (2007)
Baum, L. et al. Methylenetetrahydrofolate reductase gene A222V polymorphism and risk of ischemic stroke. Clin. Chem. Lab. Med. 42, 1370–1376 (2004)
Acknowledgements
We thank the patients and their family members whose contribution made this work possible; the nurses at Noatun (deCODE’s sample recruitment center), personnel at the deCODE core facilities, and M. Shea for the ongoing enrolment of patients at Massachusetts General Hospital; and A. Plourde and S. Makino for technical assistance.
Author Contributions D.F.G., D.O.A., A.H., S.G., P.T.E., J.R. U.T. and K.S. wrote the first draft of the paper. D.O.A., H.H., R.S., J.T.S. and G.T. collected and diagnosed the Icelandic AF samples. Ko.K. and J.H. collected and diagnosed the Swedish samples. K.L.F., S.M.G., M.S., P.K., C.A.M., E.E.S., J.R. and P.T.E. collected and diagnosed the US samples. M.C.Y.N., L.B., W.Y.S., K.S.W., J.C.N.C. and R.C.W.M collected and diagnosed the Hong Kong samples. A.H., S.G., A.S., A.J., A.B., T.B., V.M.B., G.A.H. and E.P. performed genotyping and experimental work. D.F.G., G.T., A.P., P.S., A.H. and A.K. analyzed the data. D.F.G., D.O.A., A.H., S.G., Kr.K., J.R., J.H., R.C.W.M., P.T.E, G.T, J.R.G., A.K., U.T. and K.S. planned, supervised and coordinated the work. All authors contributed to the final version of the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Some of the authors own stock and/or stock options in deCODE genetics, Inc.
Supplementary information
Supplementary information
This file contains Supplementary Methods, Supplementary Tables 1-9, Supplementary Figures 1-4 and additional references. (PDF 612 kb)
Rights and permissions
About this article
Cite this article
Gudbjartsson, D., Arnar, D., Helgadottir, A. et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 448, 353–357 (2007). https://doi.org/10.1038/nature06007
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06007
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.