Abstract
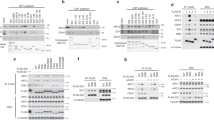
Polo kinases have crucial conserved functions in controlling the eukaryotic cell cycle through orchestrating several events during mitosis1,2. An essential element of cell cycle control is exerted by altering the expression of key regulators3. Here we show an important function for the polo kinase Cdc5p in controlling cell-cycle-dependent gene expression that is crucial for the execution of mitosis in the model eukaryote Saccharomyces cerevisiae. In particular, we find that Cdc5p is temporally recruited to promoters of the cell-cycle-regulated CLB2 gene cluster, where it targets the Mcm1p–Fkh2p–Ndd1p transcription factor complex, through direct phosphorylation of the coactivator protein Ndd1p. This phosphorylation event is required for the normal temporal expression of cell-cycle-regulated genes such as CLB2 and SWI5 in G2/M phases. Furthermore, severe defects in cell division occur in the absence of Cdc5p-mediated phosphorylation of Ndd1p. Thus, polo kinase is required for the production of key mitotic regulators, in addition to previously defined roles in controlling other mitotic events.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Barr, F. A., Silljé, H. H. W. & Nigg, E. A. Polo-like kinases and the orchestration of cell division. Nature Rev. Mol. Cell Biol. 5, 429–441 (2004)
Lee, K. S., Park, J. E., Asano, S. & Park, C. J. Yeast polo-like kinases: functionally conserved multitask mitotic regulators. Oncogene 24, 217–229 (2005)
Wittenberg, C. & Reed, S. I. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24, 2746–2755 (2005)
Alexandru, G., Uhlmann, F., Mechtler, K., Poupart, M. & Nasmyth, K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459–472 (2001)
Hu, F. et al. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107, 655–665 (2001)
Geymonat, M., Spanos, A., Walker, P. A., Johnston, L. H. & Sedgwick, S. G. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 278, 14591–14594 (2003)
Sakchaisri, K. et al. Coupling morphogenesis to mitotic entry. Proc. Natl Acad. Sci. USA 101, 4124–4129 (2004)
Futcher, B. Transcriptional regulatory networks and the yeast cell cycle. Curr. Opin. Cell Biol. 14, 676–683 (2002)
Koranda, M., Schleiffer, A., Endler, L. & Ammerer, G. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406, 94–98 (2000)
Kumar, R. et al. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10, 896–906 (2000)
Pic, A. et al. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19, 3750–3761 (2000)
Zhu, G. et al. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406, 90–94 (2000)
Breeden, L. L. Cyclin transcription: Timing is everything. Curr. Biol. 10, R586–R588 (2000)
Pic-Taylor, A., Darieva, Z., Morgan, B. A. & Sharrocks, A. D. Regulation of cell cycle-specific gene expression through cyclin-dependent kinase-mediated phosphorylation of the forkhead transcription factor Fkh2p. Mol. Cell. Biol. 24, 10036–10046 (2004)
Darieva, Z. et al. Cell cycle regulated transcription through the FHA domain of Fkh2p and the coactivator Ndd1p. Curr. Biol. 13, 1740–1745 (2003)
Reynolds, D. et al. Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev. 17, 1789–1802 (2003)
Cheng, L., Hunke, L. & Hardy, C. F. Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p. Mol. Cell. Biol. 18, 7360–7370 (1998)
Nakajima, H., Toyoshima-Morimoto, F., Taniguchi, E. & Nishida, E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 278, 25277–25280 (2003)
Song, S. & Lee, K. S. A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J. Cell Biol. 152, 451–469 (2001)
Althoefer, H., Schleiffer, A., Wassmann, K., Nordheim, A. & Ammerer, G. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae.. Mol. Cell. Biol. 15, 5917–5928 (1995)
Spellman, P. T. et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273–3297 (1998)
Simon, I. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106, 697–708 (2001)
Asano, S. et al. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 24, 2194–2204 (2005)
Zilahi, E., Salimova, E., Simanis, V. & Sipiczki, M. The S. pombe sep1 gene encodes a nuclear protein that is required for periodic expression of the cdc15 gene. FEBS Lett. 481, 105–108 (2000)
Buck, V. et al. Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J. Cell Sci. 117, 5623–5632 (2004)
Bulmer, R. et al. The forkhead transcription factor Fkh2 regulates the cell division cycle of Schizosaccharomyces pombe. Eukaryot. Cell 3, 944–954 (2004)
Rustici, G. et al. Periodic gene expression program of the fission yeast cell cycle. Nature Genet. 36, 809–817 (2004)
Alvarez, B., Martinez, A. C., Burgering, B. M. & Carrera, A. C. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413, 744–747 (2001)
Laoukili, J. et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nature Cell Biol. 7, 126–136 (2005)
Anderson, M. et al. Plo1+ regulates gene transcription at the M–G1 interval during the fission yeast mitotic cell cycle. EMBO J. 21, 5745–5755 (2002)
Acknowledgements
We thank A. Clancy for technical assistance; I. Hagan, P. March, M. Jackson and G. Pereira for advice; A. Whitmarsh, S.-H. Yang and members of our laboratories for comments on the manuscript and helpful discussions; and M. Walberg, U. Surana, G. Pereira and D. Lydall for reagents. This work was supported by Cancer Research UK, the BBSRC, the MRC and the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Methods and Supplementary Figure Legends. (DOC 75 kb)
Supplementary Figures
This file contains Supplementary Figures 1–11. (PDF 291 kb)
Supplementary Data
This is a list of the key genes and proteins used in this study. (DOC 23 kb)
Rights and permissions
About this article
Cite this article
Darieva, Z., Bulmer, R., Pic-Taylor, A. et al. Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein . Nature 444, 494–498 (2006). https://doi.org/10.1038/nature05339
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05339
This article is cited by
-
Cyclin/Forkhead-mediated coordination of cyclin waves: an autonomous oscillator rationalizing the quantitative model of Cdk control for budding yeast
npj Systems Biology and Applications (2021)
-
Forkhead transcription factor Fkh1: insights into functional regulatory domains crucial for recruitment of Sin3 histone deacetylase complex
Current Genetics (2021)
-
A Weakened Geomagnetic Field: Effects on Genomic Transcriptiln Activity, Learning, and Memory in Drosophila Melanogaster
Neuroscience and Behavioral Physiology (2018)
-
Functions and regulation of the Polo-like kinase Cdc5 in the absence and presence of DNA damage
Current Genetics (2018)
-
Degradation of Ndd1 by APC/CCdh1 generates a feed forward loop that times mitotic protein accumulation
Nature Communications (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.