Abstract
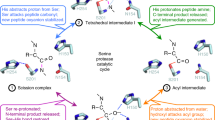
Escherichia coli GlpG is an integral membrane protein that belongs to the widespread rhomboid protease family. Rhomboid proteases, like site-2 protease (S2P) and γ-secretase, are unique in that they cleave the transmembrane domain of other membrane proteins. Here we describe the 2.1 Å resolution crystal structure of the GlpG core domain. This structure contains six transmembrane segments. Residues previously shown to be involved in catalysis, including a Ser–His dyad, and several water molecules are found at the protein interior at a depth below the membrane surface. This putative active site is accessible by substrate through a large ‘V-shaped’ opening that faces laterally towards the lipid, but is blocked by a half-submerged loop structure. These observations indicate that, in intramembrane proteolysis, the scission of peptide bonds takes place within the hydrophobic environment of the membrane bilayer. The crystal structure also suggests a gating mechanism for GlpG that controls substrate access to its hydrophilic active site.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Brown, M. S. & Goldstein, J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997)
Mattson, M. P. Pathways towards and away from Alzheimer’s disease. Nature 430, 631–639 (2004)
Sakai, J. et al. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85, 1037–1046 (1996)
Rawson, R. B. et al. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1, 47–57 (1997)
Haass, C. Take five—BACE and the γ-secretase quartet conduct Alzheimer’s amyloid β-peptide generation. EMBO J. 23, 483–488 (2004)
Levitan, D. & Greenwald, I. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer’s disease gene. Nature 377, 351–354 (1995)
De Strooper, B. et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999)
Rudner, D. Z., Fawcett, P. & Losick, R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl Acad. Sci. USA 96, 14765–14770 (1999)
Weihofen, A., Binns, K., Lemberg, M. K., Ashman, K. & Martoglio, B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296, 2215–2218 (2002)
Fluhrer, R. et al. A γ-secretase-like intramembrane cleavage of TNFα by the GxGD aspartyl protease SPPL2b. Nature Cell Biol. 8, 894–896 (2006)
Friedmann, E. et al. SPPL2a and SPPL2b promote intramembrane proteolysis of TNFα in activated dendritic cells to trigger IL-12 production. Nature Cell Biol. 8, 843–848 (2006)
Urban, S., Lee, J. R. & Freeman, M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–182 (2001)
Gallio, M., Sturgill, G., Rather, P. & Kylsten, P. A conserved mechanism for extracellular signaling in eukaryotes and prokaryotes. Proc. Natl Acad. Sci. USA 99, 12208–12213 (2002)
Wasserman, J. D., Urban, S. & Freeman, M. A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signaling. Genes Dev. 14, 1651–1663 (2000)
Koonin, E. V. et al. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 4, R19 (2003)
Urban, S., Schlieper, D. & Freeman, M. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr. Biol. 12, 1507–1512 (2002)
Sturtevant, M. A., Roark, M. & Bier, E. The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev. 7, 961–973 (1993)
McQuibban, G. A., Saurya, S. & Freeman, M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423, 537–541 (2003)
Cipolat, S. et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126, 163–175 (2006)
Brossier, F., Jewett, T. J., Sibley, L. D. & Urban, S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc. Natl Acad. Sci. USA 102, 4146–4151 (2005)
Maegawa, S., Ito, K. & Akiyama, Y. Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry 44, 13543–13552 (2005)
Brown, M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398 (2000)
Wolfe, M. S. & Kopan, R. Intramembrane proteolysis: theme and variations. Science 305, 1119–1123 (2004)
Wolfe, M. S. et al. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517 (1999)
Daley, D. O. et al. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308, 1321–1323 (2005)
Urban, S. & Wolfe, M. S. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc. Natl Acad. Sci. USA 102, 1883–1888 (2005)
Lemberg, M. K. et al. Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. EMBO J. 24, 464–472 (2005)
Russ, W. P. & Engelman, D. M. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296, 911–919 (2000)
Cleland, W. W., Frey, P. A. & Gerlt, J. A. The low barrier hydrogen bond in enzymatic catalysis. J. Biol. Chem. 273, 25529–25532 (1998)
Wei, Y. et al. A novel variant of the catalytic triad in the Streptomyces scabies esterase. Nature Struct. Biol. 2, 218–223 (1995)
Zhou, G. W., Guo, J., Huang, W., Fletterick, R. J. & Scanlan, T. S. Crystal structure of a catalytic antibody with a serine protease active site. Science 265, 1059–1064 (1994)
Paetzel, M., Dalbey, R. E. & Strynadka, N. C. Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature 396, 186–190 (1998)
Tjalsma, H. et al. Conserved serine and histidine residues are critical for activity of the ER-type signal peptidase SipW of Bacillus subtilis.. J. Biol. Chem. 275, 25102–25108 (2000)
Fersht, A. Structure and Mechanism in Protein Science: a Guide to Enzyme Catalysis and Protein Folding (W.H. Freeman, New York, 1999)
Urban, S. & Freeman, M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol. Cell 11, 1425–1434 (2003)
Ye, J., Dave, U. P., Grishin, N. V., Goldstein, J. L. & Brown, M. S. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl Acad. Sci. USA 97, 5123–5128 (2000)
Lazarov, V. K. et al. Electron microscopic structure of purified, active γ-secretase reveals an aqueous intramembrane chamber and two pores. Proc. Natl Acad. Sci. USA 103, 6889–6894 (2006)
Li, X. & Greenwald, I. Membrane topology of the C. elegans SEL-12 presenilin. Neuron 17, 101510–101521 (1996)
Li, X. & Greenwald, I. Additional evidence for an eight-transmembrane-domain topology for Caenorhabditis elegans and human presenilins. Proc. Natl Acad. Sci. USA 95, 7109–7114 (1998)
Otwinowski, Z. & Minor, W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Pape, T. & Schneider, T. R. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 (2004)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Opitz, C. et al. Intramembrane cleavage of microneme proteins at the surface of the apicomplexan parasite Toxoplasma gondii. EMBO J. 21, 1577–1585 (2002)
Zhou, X. W., Blackman, M. J., Howell, S. A. & Carruthers, V. B. Proteomic analysis of cleavage events reveals a dynamic two-step mechanism for proteolysis of a key parasite adhesive complex. Mol. Cell. Proteomics 3, 565–576 (2004)
Kraulis, P. J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Merritt, E. A. & Murphy, M. E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994)
Nicholls, A., Sharp, K. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991)
Acknowledgements
We thank V. Stojanoff, H. Robinson, A. Saxena and A. Héroux at BNL NSLS beamlines for help; T. Boggon and J. Schlessinger for sharing the crystallization robot in their laboratories; and B. Turk for sharing the fluorescence spectrometer in his laboratory. X-ray diffraction data for this study were measured at beamlines X6A, X29 and X26C of NSLS. Financial support comes principally from the US Department of Energy, and from the National Institutes of Health. This work was supported by a New Scholar Award in Aging from the Ellison Medical Foundation (to Y.H.) and a gift from the Neuroscience Education and Research Foundation (to Y.H.). Author Contributions Y.W. and Y.H. purified and characterized GlpG in various detergents. Y.W. crystallized GlpG. Y.H. and Y.W. solved the structure of GlpG and wrote the paper. Y.Z. screened the expression of many constructs and conducted some initial biochemical characterizations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The atomic coordinates of GlpG have been deposited in the Protein Data Bank (accession number 2IC8), and will be released upon publication of the paper. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
Supplementary Methods and Supplementary Figures 1–5. (PDF 574 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Zhang, Y. & Ha, Y. Crystal structure of a rhomboid family intramembrane protease. Nature 444, 179–180 (2006). https://doi.org/10.1038/nature05255
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05255
This article is cited by
-
The iRhom homology domain is indispensable for ADAM17-mediated TNFα and EGF receptor ligand release
Cellular and Molecular Life Sciences (2021)
-
Coupled Transmembrane Substrate Docking and Helical Unwinding in Intramembrane Proteolysis of Amyloid Precursor Protein
Scientific Reports (2018)
-
Embedded in the Membrane: How Lipids Confer Activity and Specificity to Intramembrane Proteases
The Journal of Membrane Biology (2018)
-
Understanding Conformational Dynamics of Complex Lipid Mixtures Relevant to Biology
The Journal of Membrane Biology (2018)
-
Structural insights into the committed step of bacterial phospholipid biosynthesis
Nature Communications (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.