Abstract
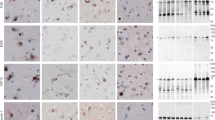
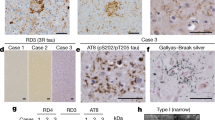
Frontotemporal dementia (FTD) is the second most common cause of dementia in people under the age of 65 years1. A large proportion of FTD patients (35–50%) have a family history of dementia, consistent with a strong genetic component to the disease2. In 1998, mutations in the gene encoding the microtubule-associated protein tau (MAPT) were shown to cause familial FTD with parkinsonism linked to chromosome 17q21 (FTDP-17)3. The neuropathology of patients with defined MAPT mutations is characterized by cytoplasmic neurofibrillary inclusions composed of hyperphosphorylated tau3,4. However, in multiple FTD families with significant evidence for linkage to the same region on chromosome 17q21 (D17S1787–D17S806), mutations in MAPT have not been found and the patients consistently lack tau-immunoreactive inclusion pathology5,6,7,8,9,10,11,12. In contrast, these patients have ubiquitin (ub)-immunoreactive neuronal cytoplasmic inclusions and characteristic lentiform ub-immunoreactive neuronal intranuclear inclusions11,12,13. Here we demonstrate that in these families, FTD is caused by mutations in progranulin (PGRN) that are likely to create null alleles. PGRN is located 1.7 Mb centromeric of MAPT on chromosome 17q21.31 and encodes a 68.5-kDa secreted growth factor involved in the regulation of multiple processes including development, wound repair and inflammation14. PGRN has also been strongly linked to tumorigenesis14. Moreover, PGRN expression is increased in activated microglia in many neurodegenerative diseases including Creutzfeldt–Jakob disease, motor neuron disease and Alzheimer's disease15,16. Our results identify mutations in PGRN as a cause of neurodegenerative disease and indicate the importance of PGRN function for neuronal survival.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Brun, A. et al. Clinical and neuropathological criteria for frontotemporal dementia. The Lund and Manchester groups. J. Neurol. Neurosurg. Psychiatry 57, 416–418 (1994)
Chow, T. W., Miller, B. L., Hayashi, V. N. & Geschwind, D. H. Inheritance of frontotemporal dementia. Arch. Neurol. 56, 817–822 (1999)
Hutton, M. et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705 (1998)
Ingram, E. M. & Spillantini, M. G. Tau gene mutations: dissecting the pathogenesis of FTDP-17. Trends Mol. Med. 8, 555–562 (2002)
Rademakers, R. et al. Tau negative frontal lobe dementia at 17q21: significant finemapping of the candidate region to a 4.8 cM interval. Mol. Psychiatry 7, 1064–1074 (2002)
Rosso, S. M. et al. Familial frontotemporal dementia with ubiquitin-positive inclusions is linked to chromosome 17q21–22. Brain 124, 1948–1957 (2001)
Lendon, C. L. et al. Hereditary dysphasic disinhibition dementia: a frontotemporal dementia linked to 17q21–22. Neurology 50, 1546–1555 (1998)
Kertesz, A. et al. Familial frontotemporal dementia with ubiquitin-positive, tau-negative inclusions. Neurology 54, 818–827 (2000)
Froelich, S. et al. Mapping of a disease locus for familial rapidly progressive frontotemporal dementia to chromosome 17q12–21. Am. J. Med. Genet. 74, 380–385 (1997)
Bird, T. D. et al. Chromosome 17 and hereditary dementia: linkage studies in three non-Alzheimer families and kindreds with late-onset FAD. Neurology 48, 949–954 (1997)
van der Zee, J. et al. A Belgian ancestral haplotype harbours a highly prevalent mutation for 17q21-linked tau-negative FTLD. Brain 129, 841–852 (2006)
Mackenzie, I. R. et al. A family with tau-negative frontotemporal dementia and neuronal intranuclear inclusions linked to chromosome 17. Brain 129, 853–867 (2006)
Rademakers, R. et al. In IPSEN Meeting Research and Perspectives in Alzheimer's Disease: Genotype–Proteotype–Phenotype Relationships in Neurodegenerative Diseases (ed. Cummings, J.) 117–137 (Springer, Paris, 2005)
He, Z. & Bateman, A. Progranulin (granulin–epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J. Mol. Med. 81, 600–612 (2003)
Malaspina, A., Kaushik, N. & de Belleroche, J. Differential expression of 14 genes in amyotrophic lateral sclerosis spinal cord detected using gridded cDNA arrays. J. Neurochem. 77, 132–145 (2001)
Baker, C. A. & Manuelidis, L. Unique inflammatory RNA profiles of microglia in Creutzfeldt–Jakob disease. Proc. Natl Acad. Sci. USA 100, 675–679 (2003)
Neary, D., Snowden, J. S. & Mann, D. M. Classification and description of frontotemporal dementias. Ann. NY Acad. Sci. 920, 46–51 (2000)
Trojanowski, J. Q. & Dickson, D. Update on the neuropathological diagnosis of frontotemporal dementias. J. Neuropathol. Exp. Neurol. 60, 1123–1126 (2001)
Mackenzie, I. R. & Feldman, H. H. Ubiquitin immunohistochemistry suggests classic motor neuron disease, motor neuron disease with dementia, and frontotemporal dementia of the motor neuron disease type represent a clinicopathologic spectrum. J. Neuropathol. Exp. Neurol. 64, 730–739 (2005)
Foster, N. L. et al. Frontotemporal dementia and parkinsonism linked to chromosome 17: a consensus conference. Ann. Neurol. 41, 706–715 (1997)
Mackenzie, I. R. & Feldman, H. Neuronal intranuclear inclusions distinguish familial FTD–MND type from sporadic cases. Acta Neuropathol. (Berl.) 105, 543–548 (2003)
Cruts, M. et al. Genomic architecture of human 17q21 linked to frontotemporal dementia uncovers a highly homologous family of low-copy repeats in the tau region. Hum. Mol. Genet. 14, 1753–1762 (2005)
Cruts, M. et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature advance online publication, doi:10.1038/nature05017 (16 July 2006)
Maquat, L. E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nature Rev. Mol. Cell Biol. 5, 89–99 (2004)
Daniel, R., He, Z., Carmichael, K. P., Halper, J. & Bateman, A. Cellular localization of gene expression for progranulin. J. Histochem. Cytochem. 48, 999–1009 (2000)
Capsoni, S. et al. Alzheimer-like neurodegeneration in aged antinerve growth factor transgenic mice. Proc. Natl Acad. Sci. USA 97, 6826–6831 (2000)
Salehi, A., Delcroix, J. D. & Swaab, D. F. Alzheimer's disease and NGF signaling. J. Neural Transm. 111, 323–345 (2004)
Lu, R. & Serrero, G. Mediation of estrogen mitogenic effect in human breast cancer MCF-7 cells by PC-cell-derived growth factor (PCDGF/granulin precursor). Proc. Natl Acad. Sci. USA 98, 142–147 (2001)
Tangkeangsirisin, W. & Serrero, G. PC cell-derived growth factor (PCDGF/GP88, progranulin) stimulates migration, invasiveness and VEGF expression in breast cancer cells. Carcinogenesis 25, 1587–1592 (2004)
Greenway, M. J. et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nature Genet. 38, 411–413 (2006)
He, Z., Ong, C. H., Halper, J. & Bateman, A. Progranulin is a mediator of the wound response. Nature Med. 9, 225–229 (2003)
Acknowledgements
We thank the FTD research team at Vancouver Coastal Health and the University of British Columbia, and particularly G. Y. R. Hsiung, for identification and follow-up of FTD families; D. Warden, P. Whitbread and E. King (OPTIMA project, Oxford, UK) for assisting with collection of UBC17 family samples; J. Chow (Department of Pathology, University of British Columbia) for help in performing the PGRN immunohistochemistry; and M. Yue, J. Gonzales (Mayo Clinic), T. de Pooter and M. Van den Broeck (University of Antwerp) for technical support. This research was funded as part of the Mayo Clinic ADRC grant from the National Institute on Aging (to M.H.), the Mayo Foundation (M.H.), and the Robert and Clarice Smith Fellowship program (to S.M.). I.R.M. and H.F. were funded by the Canadian Institutes of Health research operating grant. S.M.P.-B. received grants from the Medical Research Council (UK) and the Motor Neuron Disease Association. R.R. is a postdoctoral fellow of the Fund for Scientific Research Flanders and a visiting scientist from the Neurodegenerative Brain Diseases Group of the Department of Molecular Genetics, VIB, University of Antwerp, Belgium. Finally, we acknowledge and thank the families who contributed samples, as without them this study would not have been possible.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Methods, Supplementary Table 1 and Supplementary Figures 1–4. (PDF 2631 kb)
Rights and permissions
About this article
Cite this article
Baker, M., Mackenzie, I., Pickering-Brown, S. et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919 (2006). https://doi.org/10.1038/nature05016
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05016
This article is cited by
-
Progranulin haploinsufficiency mediates cytoplasmic TDP-43 aggregation with lysosomal abnormalities in human microglia
Journal of Neuroinflammation (2024)
-
Neuropathological hints from CSF and serum biomarkers in corticobasal syndrome (CBS): a systematic review
Neurological Research and Practice (2024)
-
Hotspots and trends of microglia in Alzheimer's disease: a bibliometric analysis during 2000–2022
European Journal of Medical Research (2024)
-
Progranulin and GPNMB: interactions in endo-lysosome function and inflammation in neurodegenerative disease
Journal of Neuroinflammation (2023)
-
TMEM106B aggregation in neurodegenerative diseases: linking genetics to function
Molecular Neurodegeneration (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.