Abstract
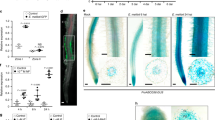
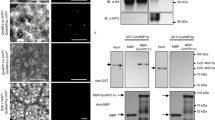
Legumes, such as Medicago truncatula, form mutualistic symbiotic relationships with nitrogen-fixing rhizobial bacteria. This occurs within specialized root organs—nodules—that provide the conditions required for nitrogen fixation. A rhizobium-derived signalling molecule, Nod factor, is required to establish the symbiosis. Perception of Nod factor in the plant leads to the induction of Ca2+ oscillations1, and the transduction of this Ca2+ signal requires DMI3 (refs 2, 3), which encodes the protein kinase Ca2+/calmodulin-dependent protein kinase (CCaMK). Central to the regulation of CCaMK is an autoinhibitory domain that negatively regulates kinase activity. Here we show that the specific removal of the autoinhibition domain leads to the autoactivation of the nodulation signalling pathway in the plant, with the resultant induction of nodules and nodulation gene expression in the absence of bacterial elicitation. This autoactivation requires nodulation-specific transcriptional regulators in the GRAS family. This work demonstrates that the release of autoinhibition from CCaMK after calmodulin binding is a central switch that is sufficient to activate nodule morphogenesis. The fact that a single regulation event is sufficient to induce nodulation highlights the possibility of transferring this process to non-legumes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ehrhardt, D. W., Wais, R. & Long, S. R. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85, 673–681 (1996)
Levy, J. et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303, 1361–1364 (2004)
Mitra, R. M. et al. A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl Acad. Sci. USA 101, 4701–4705 (2004)
Patil, S., Takezawa, D. & Poovaiah, B. W. Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium-binding domain. Proc. Natl Acad. Sci. USA 92, 4897–4901 (1995)
Takezawa, D., Ramachandiran, S., Paranjape, V. & Poovaiah, B. W. Dual regulation of a chimeric plant serine/threonine kinase by calcium and calcium/calmodulin. J. Biol. Chem. 271, 8126–8132 (1996)
Sathyanarayanan, P. V., Cremo, C. R. & Poovaiah, B. W. Plant chimeric Ca2+/calmodulin-dependent protein kinase. Role of the neural visinin-like domain in regulating autophosphorylation and calmodulin affinity. J. Biol. Chem. 275, 30417–30422 (2000)
Ramachandiran, S., Takezawa, D., Wang, W. & Poovaiah, B. W. Functional domains of plant chimeric calcium/calmodulin-dependent protein kinase: regulation by autoinhibitory and visinin-like domains. J. Biochem. 121, 984–990 (1997)
Journet, E.-P. et al. Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14, 737–748 (2001)
Tirichine, L. et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature doi:10.1038/nature04862 (this issue).
Sathyanarayanan, P. V., Siems, W. F., Jones, J. P. & Poovaiah, B. W. Calcium-stimulated autophosphorylation site of plant chimeric calcium/calmodulin-dependent protein kinase. J. Biol. Chem. 276, 32940–32947 (2001)
Ane, J. M. et al. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367 (2004)
Catoira, R. et al. Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12, 1647–1665 (2000)
Endre, G. et al. A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966 (2002)
Oldroyd, G. E. & Downie, J. A. Calcium, kinases and nodulation signalling in legumes. Nature Rev. Mol. Cell Biol. 5, 566–576 (2004)
Oldroyd, G. E. & Long, S. R. Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 131, 1027–1032 (2003)
Smit, P. et al. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791 (2005)
Kalo, P. et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789 (2005)
Boisson-Dernier, A. et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14, 695–700 (2001)
Ehrhardt, D. W., Atkinson, E. M. & Long, S. R. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256, 998–1000 (1992)
Boivin, C., Camut, S., Malpica, C. A., Truchet, G. & Rosenberg, C. Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2, 1157–1170 (1990)
Yang, T., Chaudhuri, S., Yang, L., Chen, Y. & Poovaiah, B. W. Calcium/calmodulin up-regulates a cytoplasmic receptor-like kinase in plants. J. Biol. Chem. 279, 42552–42559 (2004)
Acknowledgements
We thank D. Barker for providing the ENOD11-GUS lines; A. Downie for his critical review of the manuscript and for discussions; H. Miwa for assistance with the confocal microscope; K. Findlay and S. Bunnewell for assistance with the histological studies; and S. Long for efforts in facilitating this collaborative work. This study was supported by the BBSRC as a David Philips Fellowship to G.O. and a grant, by the Royal Society as a Wolfson Merit award to G.O., by Washington State University Agricultural Research Center and by the US National Science Foundation.Author Contributions A.M. defined conditions and generated constructs for protein expression; S.C. and T.Y. did the in-vitro biochemistry; C.G. completed all the rest of the experimental work and wrote the paper with G.O.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Methods
A complete list of the primers used to generate the DMI3 deletion constructs and a more detailed description of protocols. (DOC 29 kb)
Supplementary Table 1
ENOD11-GUS induction in dmi1, dmi2, nsp1, and nsp2 transformed with the DMI3 deletion constructs. (DOC 26 kb)
Supplementary Figure 1
Localisation of DMI3 and DMI3 deletion constructs fused to GFP (pdf 2.8 MB). (PDF 555 kb)
Supplementary Figure 2
Activation of ENOD11-GUS by DMI3-T271A and DMI3-T271D in dmi3-1 and DMI3 1-326 in dmi1 and nsp1 lines (pdf 556 KB). (PDF 2816 kb)
Rights and permissions
About this article
Cite this article
Gleason, C., Chaudhuri, S., Yang, T. et al. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441, 1149–1152 (2006). https://doi.org/10.1038/nature04812
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04812
This article is cited by
-
Exploring a rhizobium to fix nitrogen in non-leguminous plants by using a tumor-formation root pathogen
Phytopathology Research (2022)
-
Nutrient regulation of lipochitooligosaccharide recognition in plants via NSP1 and NSP2
Nature Communications (2022)
-
Calcium spikes, waves and oscillations in plant development and biotic interactions
Nature Plants (2020)
-
An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages
Nature Plants (2020)
-
Identification and expression of DoCCaMK during Sebacina sp. symbiosis of Dendrobium officinale
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.