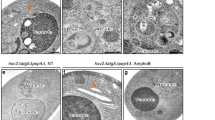
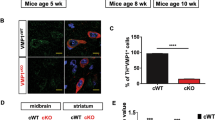
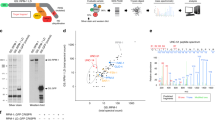
Abstract
Autophagy is an intracellular bulk degradation process through which a portion of the cytoplasm is delivered to lysosomes to be degraded1,2,3,4. Although the primary role of autophagy in many organisms is in adaptation to starvation, autophagy is also thought to be important for normal turnover of cytoplasmic contents, particularly in quiescent cells such as neurons. Autophagy may have a protective role against the development of a number of neurodegenerative diseases5,6,7,8. Here we report that loss of autophagy causes neurodegeneration even in the absence of any disease-associated mutant proteins. Mice deficient for Atg5 (autophagy-related 5) specifically in neural cells develop progressive deficits in motor function that are accompanied by the accumulation of cytoplasmic inclusion bodies in neurons. In Atg5-/- cells, diffuse, abnormal intracellular proteins accumulate, and then form aggregates and inclusions. These results suggest that the continuous clearance of diffuse cytosolic proteins through basal autophagy is important for preventing the accumulation of abnormal proteins, which can disrupt neural function and ultimately lead to neurodegeneration.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cuervo, A. M. Autophagy: in sickness and in health. Trends Cell Biol. 14, 70–77 (2004)
Levine, B. & Klionsky, D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 (2004)
Klionsky, D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118, 7–18 (2005)
Mizushima, N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 12, 1535–1541 (2005)
Ravikumar, B., Duden, R. & Rubinsztein, D. C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11, 1107–1117 (2002)
Fortun, J., Dunn, W. A. Jr, Joy, S., Li, J. & Notterpek, L. Emerging role for autophagy in the removal of aggresomes in Schwann cells. J. Neurosci. 23, 10672–10680 (2003)
Ravikumar, B. et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature Genet. 36, 585–595 (2004)
Iwata, A. et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl Acad. Sci. USA 102, 13135–13140 (2005)
Mizushima, N., Ohsumi, Y. & Yoshimori, T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 27, 421–429 (2002)
Kuma, A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004)
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005)
Betz, U. A., Vosshenrich, C. A., Rajewsky, K. & Muller, W. Bypass of lethality with mosaic mice generated by Cre–loxP-mediated recombination. Curr. Biol. 6, 1307–1316 (1996)
Mizushima, N. et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–667 (2001)
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000)
Cote, F., Collard, J. F. & Julien, J. P. Progressive neuronopathy in transgenic mice expressing the human neurofilament heavy gene: a mouse model of amyotrophic lateral sclerosis. Cell 73, 35–46 (1993)
Mangiarini, L. et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 (1996)
Kikuchi, T., Mukoyama, M., Yamazaki, K. & Moriya, H. Axonal degeneration of ascending sensory neurons in gracile axonal dystrophy mutant mouse. Acta Neuropathol. (Berl.) 80, 145–151 (1990)
Sotelo, C. Axonal abnormalities in cerebellar Purkinje cells of the ‘hyperspiny Purkinje cell’ mutant mouse. J. Neurocytol. 19, 737–755 (1990)
Lossi, L., Mioletti, S. & Merighi, A. Synapse-independent and synapse-dependent apoptosis of cerebellar granule cells in postnatal rabbits occur at two subsequent but partly overlapping developmental stages. Neuroscience 112, 509–523 (2002)
Sakai, K. & Miyazaki, J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem. Biophys. Res. Commun. 237, 318–324 (1997)
Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. Inducible gene targeting in mice. Science 269, 1427–1429 (1995)
Kopito, R. R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 (2000)
Saudou, F., Finkbeiner, S., Devys, D. & Greenberg, M. E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95, 55–66 (1998)
Kuemmerle, S. et al. Huntington aggregates may not predict neuronal death in Huntington's disease. Ann. Neurol. 46, 842–849 (1999)
Taylor, J. P. et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 12, 749–757 (2003)
Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004)
Tanaka, M. et al. Aggresomes formed by α-synuclein and synphilin-1 are cytoprotective. J. Biol. Chem. 279, 4625–4631 (2004)
Bjorkoy, G. et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 (2005)
Mori, H. et al. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nature Med. 10, 739–743 (2004)
Yamamoto, A. et al. Stacks of flattened smooth endoplasmic reticulum highly enriched in inositol 1,4,5-trisphosphate (InsP3) receptor in mouse cerebellar Purkinje cells. Cell Struct. Funct. 16, 419–432 (1991)
Acknowledgements
We thank H. Neko, M. Miwa and Y. Kabeya for technical assistance. We also thank J. Miyazaki for the donation of CAG-Cre transgenic mice, T. Yoshimori for the anti-LC3 antibody, E. Yamada for histological examination, M. Yuzaki for the rotarod analysis, and A. Kuma for discussion. We thank Z. Yue for critical reading of the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The authors thank the Yamada Science Foundation and the Cell Science Research Foundation for their financial support. Author Contributions T.H. performed most of the experiments to characterize the neuron-specific knockout mice. M.M. analysed Atg5-/- mice. K.N., Y.N., R.S.-M. and M.Y. generated Atg5flox chimaeric mice. A.Y. performed electron microscopy. K.M. and I.S. performed histological analysis. H.O. provided nestin-Cre mice and participated in manuscript preparation. N.M. conceived the experiments and generated the targeting vector. T.H. and N.M. wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1–8. (PDF 1935 kb)
Supplementary Figure Legends
This file contains text to accompany the above Supplementary Figures. (DOC 34 kb)
Rights and permissions
About this article
Cite this article
Hara, T., Nakamura, K., Matsui, M. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). https://doi.org/10.1038/nature04724
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04724
This article is cited by
-
Autophagy protein ATG-16.2 and its WD40 domain mediate the beneficial effects of inhibiting early-acting autophagy genes in C. elegans neurons
Nature Aging (2024)
-
The Multiple Roles of Autophagy in Neural Function and Diseases
Neuroscience Bulletin (2024)
-
Immunopathogenesis of viral infections in neurological autoimmune disease
BMC Neurology (2023)
-
Defective quality control autophagy in Hyperhomocysteinemia promotes ER stress and consequent neuronal apoptosis through proteotoxicity
Cell Communication and Signaling (2023)
-
Role of macrophage autophagy in postoperative pain and inflammation in mice
Journal of Neuroinflammation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.