Abstract
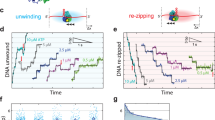
Helicases are a ubiquitous class of enzymes involved in nearly all aspects of DNA and RNA metabolism. Despite recent progress in understanding their mechanism of action, limited resolution has left inaccessible the detailed mechanisms by which these enzymes couple the rearrangement of nucleic acid structures to the binding and hydrolysis of ATP1,2. Observing individual mechanistic cycles of these motor proteins is central to understanding their cellular functions. Here we follow in real time, at a resolution of two base pairs and 20 ms, the RNA translocation and unwinding cycles of a hepatitis C virus helicase (NS3) monomer. NS3 is a representative superfamily-2 helicase essential for viral replication3, and therefore a potentially important drug target4. We show that the cyclic movement of NS3 is coordinated by ATP in discrete steps of 11 ± 3 base pairs, and that actual unwinding occurs in rapid smaller substeps of 3.6 ± 1.3 base pairs, also triggered by ATP binding, indicating that NS3 might move like an inchworm5,6. This ATP-coupling mechanism is likely to be applicable to other non-hexameric helicases involved in many essential cellular functions. The assay developed here should be useful in investigating a broad range of nucleic acid translocation motors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jankowsky, E., Gross, C. H., Shuman, S. & Pyle, A. M. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature 403, 447–451 (2000)
Lucius, A. L. & Lohman, T. M. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E. coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol. 339, 751–771 (2004)
Kolykhalov, A. A., Mihalik, K., Feinstone, S. M. & Rice, C. M. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74, 2046–2051 (2000)
Frick, D. N. Helicases as antiviral drug targets. Drug News Perspect. 16, 355–362 (2003)
Velankar, S. S., Soultanas, P., Dillingham, M. S., Subramanya, H. S. & Wigley, D. B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 (1999)
Bianco, P. R. & Kowalczykowski, S. C. Translocation step size and mechanism of the RecBC DNA helicase. Nature 405, 368–372 (2000)
Delagoutte, E. & von Hippel, P. H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: Integration of helicases into cellular processes. Q. Rev. Biophys. 36, 1–69 (2003)
Dimitrova, M., Imbert, I., Kieny, M. P. & Schuster, C. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77, 5401–5414 (2003)
Pang, P. S., Jankowsky, E., Planet, P. J. & Pyle, A. M. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 21, 1168–1176 (2002)
Levin, M. K., Gurjar, M. & Patel, S. S. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nature Struct. Mol. Biol. 12, 429–435 (2005)
Kim, J. L. et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6, 89–100 (1998)
Dohoney, K. M. & Gelles, J. Chi-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature 409, 370–374 (2001)
Bianco, P. R. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409, 374–378 (2001)
Ha, T. et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 419, 638–641 (2002)
Perkins, T. T., Li, H. W., Dalal, R. V., Gelles, J. & Block, S. M. Forward and reverse motion of single RecBCD molecules on DNA. Biophys. J. 86, 1640–1648 (2004)
Dessinges, M. N., Lionnet, T., Xi, X. G., Bensimon, D. & Croquette, V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl Acad. Sci. USA 101, 6439–6444 (2004)
Bustamante, C., Marko, J. F., Siggia, E. D. & Smith, S. Entropic elasticity of lambda-phage DNA. Science 265, 1599–1600 (1994)
Levin, M. K., Yang, Y. H. & Patel, S. S. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 279, 26005–26012 (2004)
Singleton, M. R., Dillingham, M. S., Gaudier, M., Kowalczykowski, S. C. & Wigley, D. B. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432, 187–193 (2004)
Serebrov, V. S. & Pyle, A. M. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature 430, 476–480 (2004)
Ali, J. A. & Lohman, T. M. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 276, 377–380 (1997)
Levin, M. K. & Patel, S. S. Helicase from hepatitis C virus, energetics of DNA binding. J. Biol. Chem. 277, 29377–29385 (2002)
Levin, M. K., Gurjar, M. M. & Patel, S. S. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J. Biol. Chem. 278, 23311–23316 (2003)
Tackett, A. J., Chen, Y., Cameron, C. E. & Raney, K. D. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J. Biol. Chem. 280, 10797–10806 (2005)
Tackett, A. J., Wei, L., Cameron, C. E. & Raney, K. D. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 29, 565–572 (2001)
Bianco, P. R. Hepatitis C NS3 helicase unwinds RNA in leaps and bounds. Lancet 364, 1385–1387 (2004)
Yao, N. et al. Structure of the hepatitis C virus RNA helicase domain. Nature Struct. Biol. 4, 463–467 (1997)
Smith, S. B., Cui, Y. & Bustamante, C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 361, 134–162 (2003)
Liphardt, J., Onoa, B., Smith, S. B., Tinoco, I. Jr & Bustamante, C. Reversible unfolding of single RNA molecules by mechanical force. Science 292, 733–737 (2001)
Bryant, Z. et al. Structural transitions and elasticity from torque measurements on DNA. Nature 424, 338–341 (2003)
Acknowledgements
We thank H. V. Le from the Schering-Plough Research Institute for the NS3 plasmid; S. B. Smith, P. T. X. Li, Y. R. Chemla and J.-C. Liao for discussions and technical help; T. M. Lohman for critical reading of the manuscript, and members of our laboratories for discussions and critical reading of the manuscript. This research was supported by CIHR and FQRNT doctoral fellowships (S.D.), an NIH postdoctoral fellowship (R.K.B.), NIH (I.T., A.M.P., C.B.), DOE (C.B.), and HHMI grants to investigators A.M.P. and C.B.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
Eight figures showing a variety of NS3 behaviours (Supplementary Figure 1), that the 11 bp step size is intrinsic to NS3 (Supplementary Figures 2 and 3), that two kinetic events are required for NS3 pause exit (Supplementary Figure 4), that NS3 steps consist of rapid 2–5 bp substeps (Supplementary Figure 5), and providing part of the evidence that NS3 acts as a monomer in the present assay (Supplementary Figures 6, 7 and 8). (PDF 904 kb)
Supplementary Discussion
Discussions on the variability in the step size of NS3; the comparison between bulk and single molecule experiments on both theoretical and experimental grounds; the evidence for a single active NS3 monomer; two inchworm model orientations consistent with the data; the role of ATP in coordinating translocator and helix opener movement; thermodynamic considerations on coupling between ATP and duplex unwinding; confirming substrate pairing, geometry and attachment; instrument resolution; and the effects of NS3 binding to the substrate. (PDF 188 kb)
Rights and permissions
About this article
Cite this article
Dumont, S., Cheng, W., Serebrov, V. et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439, 105–108 (2006). https://doi.org/10.1038/nature04331
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04331
This article is cited by
-
Optical tweezers in single-molecule biophysics
Nature Reviews Methods Primers (2021)
-
Kinetic and structural mechanism for DNA unwinding by a non-hexameric helicase
Nature Communications (2021)
-
Active DNA unwinding and transport by a membrane-adapted helicase nanopore
Nature Communications (2019)
-
The mechanism of DNA unwinding by the eukaryotic replicative helicase
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.