Abstract
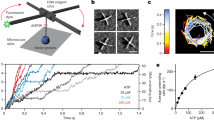
DNA gyrase is a molecular machine that uses the energy of ATP hydrolysis to introduce essential negative supercoils into DNA1,2,3. The directionality of supercoiling is ensured by chiral wrapping of the DNA4,5 around a specialized domain6,7,8,9 of the enzyme before strand passage. Here we observe the activity of gyrase in real time by tracking the rotation of a submicrometre bead attached to the side of a stretched DNA molecule10. In the presence of gyrase and ATP, we observe bursts of rotation corresponding to the processive, stepwise introduction of negative supercoils in strict multiples of two11. Changes in DNA tension have no detectable effect on supercoiling velocity, but the enzyme becomes markedly less processive as tension is increased over a range of only a few tenths of piconewtons. This behaviour is quantitatively explained by a simple mechanochemical model in which processivity depends on a kinetic competition between dissociation and rapid, tension-sensitive DNA wrapping. In a high-resolution variant of our assay, we directly detect rotational pauses corresponding to two kinetic substeps: an ATP-independent step at the end of the reaction cycle, and an ATP-binding step in the middle of the cycle, subsequent to DNA wrapping.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gellert, M., Mizuuchi, K., O'Dea, M. H. & Nash, H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA 73, 3872–3876 (1976)
Champoux, J. J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (2001)
Corbett, K. D. & Berger, J. M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33, 95–118 (2004)
Liu, L. F. & Wang, J. C. DNA–DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell 15, 979–984 (1978)
Liu, L. F. & Wang, J. C. Micrococcus luteus DNA gyrase: active components and a model for its supercoiling of DNA. Proc. Natl Acad. Sci. USA 75, 2098–2102 (1978)
Reece, R. J. & Maxwell, A. The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 19, 1399–1405 (1991)
Corbett, K. D., Shultzaberger, R. K. & Berger, J. M. The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc. Natl Acad. Sci. USA 101, 7293–7298 (2004)
Ruthenburg, A. J., Graybosch, D. M., Huetsch, J. C. & Verdine, G. L. A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J. Biol. Chem. 280, 26177–26184 (2005)
Kampranis, S. C. & Maxwell, A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl Acad. Sci. USA 93, 14416–14421 (1996)
Bryant, Z. et al. Structural transitions and elasticity from torque measurements on DNA. Nature 424, 338–341 (2003)
Brown, P. O. & Cozzarelli, N. R. A sign inversion mechanism for enzymatic supercoiling of DNA. Science 206, 1081–1083 (1979)
Levine, C., Hiasa, H. & Marians, K. J. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400, 29–43 (1998)
Wang, J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev. Mol. Cell. Biol. 3, 430–440 (2002)
Charvin, G., Strick, T. R., Bensimon, D. & Croquette, V. Tracking topoisomerase activity at the single-molecule level. Annu. Rev. Biophys. Biomol. Struct. 34, 201–219 (2005)
Bates, A. D. & Maxwell, A. DNA Topology (Oxford Univ. Press, Oxford, 2005)
Heddle, J. G., Mitelheiser, S., Maxwell, A. & Thomson, N. H. Nucleotide binding to DNA gyrase causes loss of DNA wrap. J. Mol. Biol. 337, 597–610 (2004)
Kampranis, S. C., Bates, A. D. & Maxwell, A. A model for the mechanism of strand passage by DNA gyrase. Proc. Natl Acad. Sci. USA 96, 8414–8419 (1999)
Smith, S. B., Finzi, L. & Bustamante, C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science 258, 1122–1126 (1992)
Strick, T. R., Allemand, J. F., Bensimon, D., Bensimon, A. & Croquette, V. The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 (1996)
Wang, M. D. et al. Force and velocity measured for single molecules of RNA polymerase. Science 282, 902–907 (1998)
Keller, D. & Bustamante, C. The mechanochemistry of molecular motors. Biophys. J. 78, 541–556 (2000)
Yasuda, R., Noji, H., Yoshida, M., Kinosita, K. Jr & Itoh, H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001)
Uemura, S., Higuchi, H., Olivares, A. O., De La Cruz, E. M. & Ishiwata, S. Mechanochemical coupling of two substeps in a single myosin V motor. Nature Struct. Mol. Biol. 11, 877–883 (2004)
Roca, J. & Wang, J. C. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell 71, 833–840 (1992)
Baird, C. L., Harkins, T. T., Morris, S. K. & Lindsley, J. E. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl Acad. Sci. USA 96, 13685–13690 (1999)
Pease, P. J. et al. Sequence-directed DNA translocation by purified FtsK. Science 307, 586–590 (2005)
Block, S. M. Leading the procession: new insights into kinesin motors. J. Cell Biol. 140, 1281–1284 (1998)
Vale, R. D. Myosin V motor proteins: marching stepwise towards a mechanism. J. Cell Biol. 163, 445–450 (2003)
Pato, M. L., Howe, M. M. & Higgins, N. P. A DNA gyrase-binding site at the center of the bacteriophage µ genome is required for efficient replicative transposition. Proc. Natl Acad. Sci. USA 87, 8716–8720 (1990)
Acknowledgements
We thank N. Crisona, P. Arimondo, A. Vologodskii, A. Edelstein, S. Mitelheiser, A. Schoeffler, and F. Mueller-Planitz for discussions; A. Maxwell and J. Berger for enzymes; P. Higgins for plasmids; and C. Hodges, M. Le and D. Jennings for technical assistance. J.G. acknowledges funding from the Hertz Foundation. This work was supported by the NIH and the DOE.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
A description of the function used to fit processivity and initiation rate data and a discussion of the applicability of the simple mechanochemical model from which this function is derived. (DOC 18 kb)
Supplementary Figure
A histogram of pause durations in high resolution bursts. The data support a model in which the rate-limiting step lies at the end of the reaction cycle. (DOC 88 kb)
Rights and permissions
About this article
Cite this article
Gore, J., Bryant, Z., Stone, M. et al. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature 439, 100–104 (2006). https://doi.org/10.1038/nature04319
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04319
This article is cited by
-
Light-driven high-precision cell adhesion kinetics
Light: Science & Applications (2022)
-
Rotation tracking of genome-processing enzymes using DNA origami rotors
Nature (2019)
-
Dynamic coupling between conformations and nucleotide states in DNA gyrase
Nature Chemical Biology (2018)
-
DNA supercoiling during transcription
Biophysical Reviews (2016)
-
Demonstration of virulent genes within Listeria and Klebsiella isolates contaminating the export quality frozen shrimps
International Aquatic Research (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.