Abstract
The influenza A viral heterotrimeric polymerase complex (PA, PB1, PB2) is known to be involved in many aspects of viral replication and to interact with host factors1, thereby having a role in host specificity2,3. The polymerase protein sequences from the 1918 human influenza virus differ from avian consensus sequences at only a small number of amino acids, consistent with the hypothesis that they were derived from an avian source shortly before the pandemic. However, when compared to avian sequences, the nucleotide sequences of the 1918 polymerase genes have more synonymous differences than expected, suggesting evolutionary distance from known avian strains. Here we present sequence and phylogenetic analyses of the complete genome of the 1918 influenza virus4,5,6,7,8, and propose that the 1918 virus was not a reassortant virus (like those of the 1957 and 1968 pandemics9,10), but more likely an entirely avian-like virus that adapted to humans. These data support prior phylogenetic studies suggesting that the 1918 virus was derived from an avian source11. A total of ten amino acid changes in the polymerase proteins consistently differentiate the 1918 and subsequent human influenza virus sequences from avian virus sequences. Notably, a number of the same changes have been found in recently circulating, highly pathogenic H5N1 viruses that have caused illness and death in humans and are feared to be the precursors of a new influenza pandemic. The sequence changes identified here may be important in the adaptation of influenza viruses to humans.
Similar content being viewed by others
Main
Influenza A viruses cause annual outbreaks in humans and domestic animals. Periodically, new strains emerge in humans that cause global pandemics. The severe ‘Spanish’ influenza pandemic of 1918–1919 infected hundreds of millions, and resulted in the death of approximately 50 million people12. We have previously used phylogenetic analyses to help understand the origin of the pandemic virus8,11; functional studies to understand the pathogenicity of the 1918 virus are underway6,13,14,15,16,17. Recent data have shown that viral constructs bearing the 1918 haemagglutinin gene are pathogenic in a mouse model, but the genetic basis of this observation has not yet been mapped6,13,14,15,16,17. The overall goals of this project have been to understand the origin and unusual virulence of the 1918 influenza virus.
The influenza virus A polymerase functions as a heterotrimer formed by the PB2, PB1 and PA proteins (see ref. 1 for a review). An additional small open reading frame has recently been identified, coding for a peptide (PB1-F2) that is thought to play a role in virus-induced cell death18. It is not yet clear how the polymerase complex must change to adapt to a new host3. A single amino acid change in PB2, E627K, was shown (1) to be important for mammalian adaptation2,3, (2) to distinguish highly pathogenic avian influenza (HPAI) H5N1 viruses in mice19, and (3) to be present in the single fatal human infection during the HPAI H7N7 outbreak in the Netherlands in 2003 (ref. 20), and in some recent H5N1 isolates from humans in Vietnam and Thailand and wild birds in China21,22,23.
The open reading frame sequences of segment 1 (PB2), segment 2 (PB1) and segment 3 (PA) of A/Brevig Mission/1/1918, and theoretical translations of the four identified reading frames, are shown in Supplementary Fig. 1a–c. The 1918 PB2 protein contained five changes from the avian consensus sequence (Table 1). Of these, A199S is in the area mapped as the PB1 binding site, and the L475M change is in a nuclear localization signal24,25,26. Three other changes at residues 567, 627 and 702 occur at sites that are not in known functional domains.
The 1918 PB1 protein differed from the avian consensus by seven residues (one of which is shown in Table 1; see also Supplementary Fig. 2). Of these, K54R is in the overlapping binding domains for complementary (c)RNA and viral (v)RNA. Changes at residues 375, 383 and 473 all occur in between the four conserved polymerase motifs in the cRNA binding domain27, and changes at residues 576, 645 and 654 occur in the vRNA binding domain28.
Seven changes were noted in the 1918 PA protein compared with the avian consensus (four of which are shown in Table 1, the other three being C241Y, K312R and I322V). The C241Y change occurs in a nuclear localization signal, but the other six changes (at residues 55, 100, 312, 322, 382 and 552) occur at sites outside of known functional domains24,25,26.
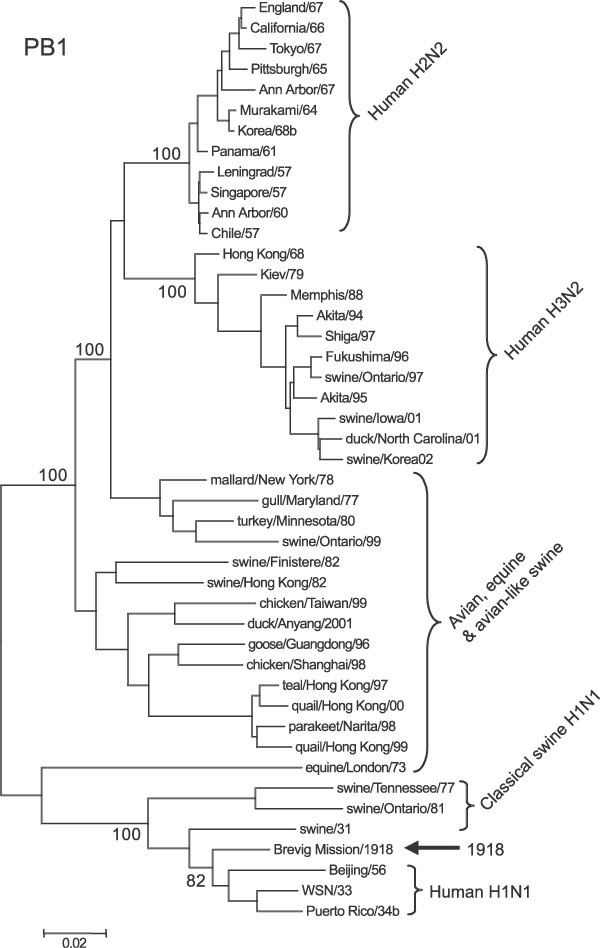
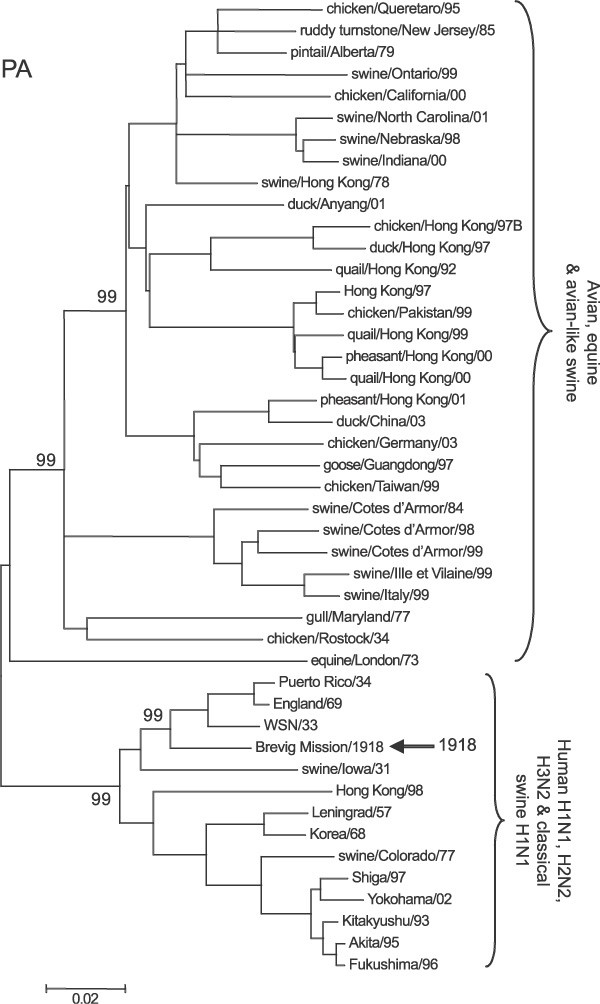
Representative phylogenetic analyses of the three polymerase genes are shown in Figs 1 –3. The 1918 human pandemic viral polymerase genes were compared to representative avian influenza genes with regards to transition/transversion (Ti/Tv) ratio, synonymous/non-synonymous (S/N) ratio, and the numbers of differences at fourfold degenerate sites (defined in ref. 11). Ti/Tv ratios for most comparisons using the 1918 viral genes and representative sequences of either North American or Eurasian avian genes yielded values between 2 and 4. This range was similar to that observed for comparisons of various avian genes with one another, except for the PB1 gene. For PB1, comparisons of the 1918 viral gene with avian virus PB1 genes was always close to 2, whereas comparisons of various avian genes with one another were in the range of 6–10. There were fewer transversions in comparisons between avian PB1 genes than in comparisons between avian PB1 and 1918 human virus PB1, probably reflecting that transversions more often lead to non-synonymous changes.
Sequences were aligned and analysed for phylogenetic relationships using the NJ algorithm, with the proportion of sequence differences as the distance measure. Bootstrap values (100 replications) for key nodes are shown (for clarity, identical and nearly identical sequences have been removed from the trees). Major clades are identified with large brackets. The arrow identifies the position of the 1918 PB2 gene sequence. A distance bar is shown below the tree. Influenza strain abbreviations used in the analyses are listed in Supplementary Table 1.
S/N ratios for most comparisons using the 1918 viral genes and representative sequences of either North American or Eurasian avian genes usually yielded values in the range of 7–16 for both the PA and PB2 genes, as is the case for most avian versus avian PA and PB2 gene comparisons. Like the Ti/Tv ratios, the S/N ratios were somewhat higher with the PB1 gene (most of the comparisons yielded ratios in the range of 16–25), owing to a smaller number of non-synonymous changes in comparisons of avian PB1 genes with one another. These findings may reflect a more conservative evolution of PB1 in birds.
A subset of synonymous differences occurs at sites that are fourfold degenerate (that is, where a substitution with any base does not result in an amino acid replacement). As these sites are not subject to selective pressure at the protein level, base substitutions at many fourfold degenerate sites may accumulate rapidly. If influenza virus genes have been evolving in birds for long enough to reach evolutionary stasis, as is suggested by the high S/N ratios described above, one would predict that at many of the sites where fourfold degeneracy is possible, all four bases would be present in the avian clade unless the constraints of RNA secondary structure limit the accumulation of synonymous changes. In fact, when avian sequences from geographically distinct lineages (North American versus European) were compared, the per cent difference at fourfold degenerate sites yielded values in the 27–38% range. In contrast, calculating the per cent difference at fourfold degenerate sites in comparisons of the 1918 viral PA, PB1 and PB2 gene sequences with avian sequences yielded consistently higher values (range 41–51%) for all three genes. As with the other 1918 genes11, this suggests that the donor source of the 1918 virus was in evolutionary isolation from those avian influenza viruses currently represented in the databases.
Emphasizing the avian-like nature of the 1918 influenza virus polymerase proteins, out of 19 total amino acid changes from the avian consensus, there are only 10 amino acid positions (out of 2,232 total codons) that consistently distinguish the 1918 and subsequent human polymerase proteins PB2, PB1 and PA from their avian influenza counterparts (these are defined as changes from avian sequences in the 1918 virus that are maintained without change in subsequent human viruses) (Table 1). It is likely that these changes have an important role in human adaptation. Seven of these ten changes were previously noted in an alignment between avian and human influenza polymerases3. What follows is a comparison between the 1918 virus changes and recent H5N1 isolates, in order to evaluate possible examples of parallel evolution in the adaptation of avian influenza viruses to humans.
In the PB2 protein, five changes distinguish the human isolates from avian sequences (Table 1). Out of 253 available PB2 sequences from human H1N1, H2N2 and H3N2 isolates, these five changes are almost completely preserved, with the exception that two recent H3N2 isolates have the avian Lys residue at position 702. Only a small number of avian influenza isolates show any of these five changes, and it is intriguing that almost all of these isolates are from HPAI H5N1 or H7N7 viruses, or from the H9N2 lineage that infected a small number of humans in China in the late 1990s (ref. 29). Only 5 out of 282 available avian PB2 sequences have a Ser residue at position 199, four of these being 1997 H5N1 isolates from Hong Kong. The A199S change was also found in 5 out of 18 H5N1 strains isolated from humans (all five were from the 1997 Hong Kong outbreak). Of the avian viruses, 36 out of 336 have an Arg residue at position 702, 30 of which are H9N2 isolates from China around 1996–2000, and 5 are H5N1 isolates from Hong Kong in 1997 and 2001. Out of 18 available 1997 H5N1 strains isolated from humans, three have the K702R change.
Perhaps most interestingly, the 1918 virus and subsequent human isolates have a Lys residue at position 627. This residue has been implicated in host adaptation2,3, and has previously been shown to be crucial for high pathogenicity in mice infected with the 1997 H5N1 virus19. Of the avian isolates, 19 out of 345 have a Lys residue at position 627, 18 of which are HPAI H5N1 or H7N7 avian influenza viruses. Sixteen of these were recently characterized H5N1 isolates from a die-off of wild waterfowl around Qinghai Lake in western China in 2005 (ref. 21). In human H5N1 isolates, 11 out of 37 have the E627K change: A/Hong Kong/483/1997 and A/Hong Kong/485/1997, four out of six isolates from Vietnam in 2004 (ref. 22), and two out of three isolates from Thailand in 2004 (ref. 23). The E627K mutation was seen in six out of seven H5N1 isolates from Thai tigers in 2004, and was also present in the H7N7 virus responsible for the single human fatality during the HPAI H7N7 outbreak in the Netherlands in 2003 (ref. 20). It was not noted in the contemporaneous chicken isolates.
At position 475, only one out of 355 avian isolates has a Met residue (an H5N1 HPAI virus from 2004). Similarly, only one out of 345 avian viruses has an Asn residue at position 567. None of the human H5N1 isolates has the L475M or the D567N changes. None of the available H5N1 or H7N7 sequences has more than one of the proposed human-adaptive PB2 changes determined for the 1918 virus.
The PA protein shows a similar pattern: four residues consistently differ between 1918 and subsequent human isolates and the avian consensus sequence (Table 1). Three other changes (C241Y, K312R and I322V) distinguish 1918, H1N1 and H2N2 human isolates, but most H3N2 isolates have the avian amino acid at these positions. Of 295 available sequences from human H1N1, H2N2 and H3N2 isolates, all have Asn at position 55 (except A/WSN/33), Ala at position 100 and Ser at position 552. Only 5 out of 295 human isolates have the avian Glu residue at position 382. Notably, these five isolates make up a minor clade of recent H3N2 isolates that have a number of unusual changes from typical human H3N2 viruses30. When avian influenza sequences are analysed, none (out of 209 sequences) has Asn at position 55 or Ser at position 552. Only 8 out of 209 avian PA protein sequences show the V100A change: six recent H6N2 isolates from chickens in California, and two HPAI 2002 H5N1 duck isolates from China. Of the 209 avian sequences, five have an Asp residue at position 382, including two HPAI H5N2 isolates from chickens in Mexico in 1994.
The PB1 gene segment was replaced by reassortment in both the 1957 and 1968 pandemics9. We compared the PB1 protein from the 1918 human virus with those of the avian-derived PB1 segments from the 1957 and 1968 pandemics. Human H1N1, H2N2 and H3N2 viruses derived from the 1918, 1957 and 1968 pandemics, respectively, each possessed a uniquely derived avian-like PB1 gene segment, and so we sought to identify any parallel changes that might shed light on human adaptation. The three human pandemic PB1 proteins differ from the avian consensus by only 4–7 residues each (Supplementary Fig. 2). Only one of these changes is shared among the pandemic isolates: an N375S change. This change to a serine residue is also found in swine and equine influenza A isolates. With few exceptions, all human influenza PB1 proteins have Ser at this site. Of 230 human influenza sequences, only two H1N1 isolates (A/FM/47 and A/Beijing/1956) and the ‘minor clade’ H3N2 isolates described above have the avian Asn residue30. In contrast, although this residue is maintained in almost all mammalian isolates, it is variable among avian PB1 proteins. Of 293 avian isolates, 66% have the consensus Asn residue at position 375, 18% have a Ser residue and 12% have a Thr residue.
The data presented here highlight the marked conservation of the PB1 protein in avian influenza viruses. PB1 functions as an RNA-dependent RNA polymerase, and so it is reasonable to hypothesize that its enzymatic function is optimal in this conserved form. In humans, the PB1 proteins experience linear change over time. Indeed, PB1 in humans acquires ∼0.4 amino acid changes per year. As there is such strong antigenic selection on human viruses, it is possible that although the observed changes in PB1 are selectively beneficial with respect to antigenicity, they are mildly deleterious to enzyme function. Such complex fitness trade-offs are thought to be commonplace in RNA virus evolution. Supporting this hypothesis, a recent study examining combinations of avian and human influenza polymerases showed that the most efficient influenza transcriptional activity in vitro was seen with an avian-derived PB1, even if the PB2, PA and NP proteins were from a human virus3. Acquiring an avian PB1 by reassortment might provide a replicative advantage to the new virus, possibly explaining why both of the last two pandemics and the 1918 influenza virus all had very avian-like PB1 proteins.
Both the 1957 and 1968 pandemic influenza viruses were avian/human reassortants in which 2–3 avian gene segments were reassorted with the then-circulating, human-adapted virus9,10. Unlike the 1957 and 1968 pandemics, however, the 1918 virus was most likely not a human/avian reassortant virus, but rather an avian-like virus that adapted to humans in toto8,11. On the basis of amino acid replacement rates in human influenza virus polymerase genes, it is possible that these segments were circulating in human influenza viruses as early as 1900. However, proof that the 1918 virus did not retain gene segments from the previously circulating human influenza A strain would require discovery of a sample of the pre-1918 virus from archival material. The donor source, although avian-like at the protein level, may have come from a subset of avian influenza viruses not currently represented in the sequence databases and may have been in evolutionary isolation.
The fact that amino acid changes identified in the 1918 analysis are also seen in HPAI strains of H5N1 and H7N7 avian viruses that have caused fatalities in humans is intriguing, and suggests that these changes may facilitate virus replication in human cells and increase pathogenicity. It is possible that the high pathogencity of the 1918 virus was related to its emergence as a human-adapted avian influenza virus. These changes may reflect a process of parallel evolution as avian influenza A viruses mutate in response to adaptational pressures, and suggest that the genetic basis of avian influenza virus adaptation to humans can be mapped.
Methods
RNA isolation, amplification and sequencing
RNA was isolated from frozen 1918 human lung tissue using Trizol (Invitrogen) according to the manufacturer's instructions. Each fragment was reverse transcribed, amplified, and sequenced at least twice. Reverse transcription polymerase chain reaction (RT–PCR), isolation of products and sequencing have been previously described4. Lists of primers and primer sequences are available upon request. Replicate RT–PCR reactions from independently produced RNA preparations gave identical sequence results. The 2,280-nucleotide complete coding sequence of PB2 was amplified in 33 overlapping fragments. The 2,274-nucleotide coding sequence of PB1 was amplified in 33 overlapping fragments. The 2,151-nucleotide coding sequence of PA was amplified in 32 overlapping fragments. The PCR products ranged in size from 77–138 bp.
Phylogenetic analyses
Phylogenetic analyses of the three polymerase genes were done using standard methods. We generated trees using the neighbour-joining (NJ) algorithm, with proportion of differences as the distance measure using MEGA version 2.1. Character evolution was analysed with the MacClade program after a parsimony analysis using PAUP version 4.0 beta, using ACTRAN as the optimization method. Trees were also generated using maximum-likelihood with midpoint rooting. All algorithms generated comparable trees, with major clades representing human, classical swine and avian-like viruses (NJ trees shown in Figs 1 –3; complete data set available upon request). Polymerase segment sequences used in this analysis were obtained from GenBank and the Influenza Sequence Databank (ISD). (See Supplementary Table 1 for a list of sequences used.) For the PB2 gene, 83 sequences were used, all of which were full length. For the PB1 gene, 91 sequences were used, three of which were not full length. For the PA gene, 105 sequences were used, six of which were not full length.
References
Fodor, E. & Brownlee, G. G. in Influenza (ed. Potter, C. W.) 1–29 (Elsevier, Amsterdam, 2002)
Subbarao, E. K., London, W. & Murphy, B. R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67, 1761–1764 (1993)
Naffakh, N., Massin, P., Escriou, N., Crescenzo-Chaigne, B. & van der Werf, S. Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J. Gen. Virol. 81, 1283–1291 (2000)
Reid, A. H., Fanning, T. G., Hultin, J. V. & Taubenberger, J. K. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl Acad. Sci. USA 96, 1651–1656 (1999)
Reid, A. H., Fanning, T. G., Janczewski, T. A. & Taubenberger, J. K. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc. Natl Acad. Sci. USA 97, 6785–6790 (2000)
Basler, C. F. et al. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl Acad. Sci. USA 98, 2746–2751 (2001)
Reid, A. H., Fanning, T. G., Janczewski, T. A., McCall, S. & Taubenberger, J. K. Characterization of the 1918 “Spanish” influenza virus matrix gene segment. J. Virol. 76, 10717–10723 (2002)
Reid, A. H., Fanning, T. G., Janczewski, T. A., Lourens, R. & Taubenberger, J. K. Novel origin of the 1918 pandemic influenza virus nucleoprotein gene segment. J. Virol. 78, 12462–12470 (2004)
Kawaoka, Y., Krauss, S. & Webster, R. G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63, 4603–4608 (1989)
Scholtissek, C., Rohde, W., Von Hoyningen, V. & Rott, R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87, 13–20 (1978)
Reid, A. H., Taubenberger, J. K. & Fanning, T. G. Evidence of an absence: the genetic origins of the 1918 pandemic influenza virus. Nature Rev. Microbiol. 2, 909–914 (2004)
Johnson, N. P. & Mueller, J. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76, 105–115 (2002)
Geiss, G. K. et al. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl Acad. Sci. USA 99, 10736–10741 (2002)
Tumpey, T. M. et al. Existing antivirals are effective against influenza viruses with genes from the 1918 pandemic virus. Proc. Natl Acad. Sci. USA 99, 13849–13854 (2002)
Tumpey, T. M. et al. Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proc. Natl Acad. Sci. USA 101, 3166–3171 (2004)
Kash, J. C. et al. The global host immune response: contribution of HA and NA genes from the 1918 Spanish influenza to viral pathogenesis. J. Virol. 78, 9499–9511 (2004)
Kobasa, D. et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431, 703–707 (2004)
Chen, W. et al. A novel influenza A virus mitochondrial protein that induces cell death. Nature Med. 7, 1306–1312 (2001)
Shinya, K. et al. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 320, 258–266 (2004)
Fouchier, R. A. et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl Acad. Sci. USA 101, 1356–1361 (2004)
Chen, H. et al. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436, 191–192 (2005)
Li, K. S. et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430, 209–213 (2004)
Puthavathana, P. et al. Molecular characterization of the complete genome of human influenza H5N1 virus isolates from Thailand. J. Gen. Virol. 86, 423–433 (2005)
Toyoda, T., Adyshev, D. M., Kobayashi, M., Iwata, A. & Ishihama, A. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit–subunit contact sites. J. Gen. Virol. 77, 2149–2157 (1996)
Masunaga, K., Mizumoto, K., Kato, H., Ishihama, A. & Toyoda, T. Molecular mapping of influenza virus RNA polymerase by site-specific antibodies. Virology 256, 130–141 (1999)
Ohtsu, Y., Honda, Y., Sakata, Y., Kato, H. & Toyoda, T. Fine mapping of the subunit binding sites of influenza virus RNA polymerase. Microbiol. Immunol. 46, 167–175 (2002)
Biswas, S. K. & Nayak, D. P. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68, 1819–1826 (1994)
Gonzalez, S. & Ortin, J. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 18, 3767–3775 (1999)
Guo, Y. J. et al. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 267, 279–288 (2000)
Holmes, E. C. et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 3, e300 (2005)
Acknowledgements
The research described in this report was done using stringent safety precautions to protect the laboratory workers, the environment and the public from this virus. The intention of this research is to provide the basis for understanding how influenza pandemic strains form and to help ascertain the risk of future influenza pandemics. This study was partially supported by a grant to J.K.T. from the National Institutes of Health, and by intramural funds from the Armed Forces Institute of Pathology. The opinions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the US Department of the Army or the US Department of Defense.Author Contributions J.K.T. planned the project, and A.H.R., R.M.L., R.W. and G.J. generated the sequence data. J.K.T., A.H.R. and T.G.F. performed data analysis. J.K.T. wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Coding sequences of the PB2, PB1 and PA genes have been deposited in GenBank under accession numbers DQ208309, DQ208310 and DQ208311, respectively. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Table
Influenza sequences used in the analysis (DOC 454 kb)
Supplementary Figure 1
Complete coding sequence of the 1918 influenza virus PB2 gene segment (DOC 67 kb)
Supplementary Figure 2
Theoretical translation of the 1918 influenza virus PB1 open reading frame (a) and the PB1-F2 open reading frame (b) as aligned to representative PB1 proteins from other human and animal influenza A viruses. (PPT 46 kb)
Supplementary Figure Legends
Text to accompany the above Supplementary Figures. (DOC 23 kb)
Rights and permissions
About this article
Cite this article
Taubenberger, J., Reid, A., Lourens, R. et al. Characterization of the 1918 influenza virus polymerase genes. Nature 437, 889–893 (2005). https://doi.org/10.1038/nature04230
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04230
This article is cited by
-
Are pigs overestimated as a source of zoonotic influenza viruses?
Porcine Health Management (2022)
-
Cross-species virus transmission and its pandemic potential
Bulletin of the National Research Centre (2022)
-
100-year-old pandemic flu viruses yield new genomes
Nature (2022)
-
Host adaptive mutations in the 2009 H1N1 pandemic influenza A virus PA gene regulate translation efficiency of viral mRNAs via GRSF1
Communications Biology (2022)
-
An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates
Virus Genes (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.