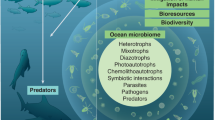
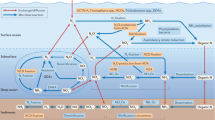
Abstract
The global ocean is an integrated living system where energy and matter transformations are governed by interdependent physical, chemical and biotic processes. Although the fundamentals of ocean physics and chemistry are well established, comprehensive approaches to describing and interpreting oceanic microbial diversity and processes are only now emerging. In particular, the application of genomics to problems in microbial oceanography is significantly expanding our understanding of marine microbial evolution, metabolism and ecology. Integration of these new genome-enabled insights into the broader framework of ocean science represents one of the great contemporary challenges for microbial oceanographers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fuhrman, J. A. & Azam, F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters. Mar. Biol. 66, 109–120 (1982).
Rappé, M. S., Connon, S. A., Vergin, K. L. & Giovannoni, S. J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633 (2002).
Olsen, G. J., Lane, D. J., Giovannoni, S. J., Pace, N. R. & Stahl, D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40, 337–365 (1986).
DeLong, E. F., Wickham, G. S. & Pace, N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243, 1360–1363 (1989).
Stein, J. L., Marsh, T. L., Wu, K. Y., Shizuya, H. & DeLong, E. F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178, 591–599 (1996).
Handelsman, J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685 (2004).
Karl, D. M. Cellular nucleotide measurements and applications in microbial ecology. Microbiol. Rev. 44, 739–796 (1980).
Hobbie, J. E., Daley, R. J. & Jasper, S. Use of Nuclepore™ filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33, 1225–1228 (1977).
Karl, D. M. Measurement of microbial activity and growth in the ocean by rates of stable ribonucleic acid synthesis. Appl. Environ. Microbiol. 38, 850–860 (1979).
Azam, F. et al. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263 (1983).
Pomeroy, L. R. The ocean's food web, a changing paradigm. BioScience 24, 499–504 (1974).
Waterbury, J. B., Watson, S. W., Guillard, R. R. L. & Brand, L. E. Widespread ocurrence of a unicellular marine, planktonic, cyanobacterium. Nature 277, 293–294 (1979).
Johnson, P. W. & Sieburth, J. M. Chroococcoid cyanobacteria in the sea: a ubiquitous and diverse phototrophic biomass. Limnol. Oceanogr. 24, 928–935 (1979).
Chisholm, S. W. et al. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334, 340–343 (1988).
Corliss, J. B. et al. Submarine thermal springs on the Galapagos Rift. Science 203, 1073–1083 (1979).
Yayanos, A. A., Dietz, A. S. & Van Boxtel, R. Obligately barophilic bacterium from the Mariana Trench. Proc. Natl Acad. Sci. USA 78, 5212–5215 (1981).
Stahl, D. A., Lane, D. J., Olsen, G. J. & Pace, N. R. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science 224, 409–411 (1984).
Giovannoni, S. J., Britschgi, T. B., Moyer, C. L. & Field, K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345, 60–63 (1990).
DeLong, E. F. Archaea in coastal marine environments. Proc. Natl Acad. Sci. USA 89, 5685–5689 (1992).
Karner, M. B., DeLong, E. F. & Karl, D. M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510 (2001).
Fuhrman, J. A., McCallum, K. & Davis, A. A. Novel major archaebacterial group from marine plankton. Nature 356, 148–149 (1992).
Moon-van der Staay, S. Y., De Wachter, R. & Vaulot, D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409, 607–610 (2001).
Kolber, Z. S., Van Dover, C. L., Niederman, R. A. & Falkowski, P. G. Bacterial photosynthesis in surface waters of the open ocean. Nature 407, 177–179 (2000).
Béjà, O. et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902–1906 (2000).
Bergh, O., Borsheim, K. Y., Bratbak, G. & Heldal, M. High abundance of viruses found in aquatic environments. Nature 340, 467–468 (1989).
Paul, J. H., Sullivan, M. B., Segall, A. M. & Rohwer, F. Marine phage genomics. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 133, 463–476 (2002).
Falkowski, P. G. et al. The evolution of modern eukaryotic phytoplankton. Science 305, 354–360 (2004).
Lerat, E., Daubin, V., Ochman, H. & Moran, N. A. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 3, e130 (2005).
DeLong, E. F. Microbial community genomics in the ocean. Nature Rev. Microbiol. 3, 459–469 (2005).
Thompson, J. R. et al. Genotypic diversity within a natural coastal bacterioplankton population. Science 307, 1311–1313 (2005).
Armbrust, E. V. et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86 (2004).
Moran, M. A. et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432, 910–913 (2004).
Rocap, G. et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047 (2003).
Dufresne, A. et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl Acad. Sci. USA 100, 10020–10025 (2003).
Palenik, B. et al. The genome of a motile marine Synechococcus. Nature 424, 1037–1042 (2003).
Glockner, F. O. et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl Acad. Sci. USA 100, 8298–8303 (2003).
Hou, S. et al. Genome sequence of the deep-sea gammaproteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy. Proc. Natl Acad. Sci. USA 101, 18036–18041 (2004).
Rabus, R. et al. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 6, 887–902 (2004).
Ruby, E. G. et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl Acad. Sci. USA 102, 3004–3009 (2005).
Vezzi, A. et al. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307, 1459–1461 (2005).
Giovanonni, S. J. et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245 (2005).
Schmidt, T. M., DeLong, E. F. & Pace, N. R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 173, 4371–4378 (1991).
Béjà, O., Spudich, E. N., Spudich, J. L., Leclerc, M. & DeLong, E. F. Proteorhodopsin phototrophy in the ocean. Nature 411, 786–789 (2001).
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004).
Tringe, S. G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005).
Suzuki, M. T., Preston, C. M., Chavez, F. & DeLong, E. F. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in the Monterey Bay. Aquat. Microb. Ecol. 24, 117–127 (2001).
Doney, S. C., Abbott, M. R., Cullen, J. J., Karl, D. M. & Rothstein, L. From genes to ecosystems: the ocean's new frontier. Front. Ecol. Environ. 2, 457–466 (2004).
Stahl, D. A. & Tiedje, J. M. Microbial Ecology and Genomics: A Crossroads of Opportunity (American Society for Microbiology, Washington D.C., 2002).
Karl, D. M., Bidigare, R. R. & Letelier, R. M. Long-term changes in plankton community structure and productivity in the North Pacific Subtropical Gyre: The domain shift hypothesis. Deep-Sea Res. II 48, 1449–1470 (2001).
Hallam, S. J. et al. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305, 1457–1462 (2004).
Karl, D. M. Accurate estimation of microbial loop processes and rates. Microbiol. Ecol 28, 147–150 (1994).
Acknowledgements
The authors' work is supported by the NSF, the Gordon and Betty Moore Foundation, and the Department of Energy. We thank our colleagues, students and CMORE collaborators for their ideas, inspiration and enthusiasm.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
Rights and permissions
About this article
Cite this article
DeLong, E., Karl, D. Genomic perspectives in microbial oceanography. Nature 437, 336–342 (2005). https://doi.org/10.1038/nature04157
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04157
This article is cited by
-
Linking bacterial and archaeal community dynamics to related hydrological, geochemical and environmental characteristics between surface water and groundwater in a karstic estuary
Acta Oceanologica Sinica (2023)
-
A miniaturized bionic ocean-battery mimicking the structure of marine microbial ecosystems
Nature Communications (2022)
-
Priorities for ocean microbiome research
Nature Microbiology (2022)
-
The composition of cultivable bacteria, bacterial pollution, and environmental variables of the coastal areas: an example from the Southeastern Black Sea, Turkey
Environmental Monitoring and Assessment (2020)
-
Coupling virio- and bacterioplankton populations with environmental variable changes in the Bohai Sea
Acta Oceanologica Sinica (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.