Abstract
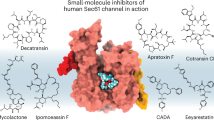
The segregation of secretory and membrane proteins to the mammalian endoplasmic reticulum is mediated by remarkably diverse signal sequences that have little or no homology with each other1,2. Despite such sequence diversity, these signals are all recognized and interpreted by a highly conserved protein-conducting channel composed of the Sec61 complex3,4. Signal recognition by Sec61 is essential for productive insertion of the nascent polypeptide into the translocation site5, channel gating6 and initiation of transport. Although subtle differences in these steps can be detected between different substrates7,8, it is not known whether they can be exploited to modulate protein translocation selectively. Here we describe cotransin, a small molecule that inhibits protein translocation into the endoplasmic reticulum. Cotransin acts in a signal-sequence-discriminatory manner to prevent the stable insertion of select nascent chains into the Sec61 translocation channel. Thus, the range of substrates accommodated by the channel can be specifically and reversibly modulated by a cell-permeable small molecule that alters the interaction between signal sequences and the Sec61 complex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
von Heijne, G. Signal sequences. The limits of variation. J. Mol. Biol. 184, 99–105 (1985)
Randall, L. L. & Hardy, S. J. Unity in function in the absence of consensus in sequence: role of leader peptides in export. Science 243, 1156–1159 (1989)
Rapoport, T. A., Jungnickel, B. & Kutay, U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 65, 271–303 (1996)
Johnson, A. E. & van Waes, M. A. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15, 799–842 (1999)
Jungnickel, B. & Rapoport, T. A. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell 82, 261–270 (1995)
Crowley, K. S., Liao, S., Worrell, V. E., Reinhart, G. D. & Johnson, A. E. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell 78, 461–471 (1994)
Rutkowski, D. T., Lingappa, V. R. & Hegde, R. S. Substrate-specific regulation of the ribosome–translocon junction by N-terminal signal sequences. Proc. Natl Acad. Sci. USA 98, 7823–7828 (2001)
Kim, S. J., Mitra, D., Salerno, J. R. & Hegde, R. S. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev. Cell 2, 207–217 (2002)
Foster, C. A. et al. Pharmacological modulation of endothelial cell-associated adhesion molecule expression: implications for future treatment of dermatological diseases. J. Dermatol. 21, 847–854 (1994)
Chen, Y., Bilban, M., Foster, C. A. & Boger, D. L. Solution-phase parallel synthesis of a pharmacophore library of HUN-7293 analogues: a general chemical mutagenesis approach to defining structure-function properties of naturally occurring cyclic (depsi)peptides. J. Am. Chem. Soc. 124, 5431–5440 (2002)
Kunkel, E. J. et al. Rapid structure-activity and selectivity analysis of kinase inhibitors by BioMAP analysis in complex human primary cell-based models. Assay Drug Dev. Technol. 2, 431–441 (2004)
Lindley, I. J. D., Marquardt-Gehrke, K. F. & Dascher-Nadel, C. Screening assay for cotranslational translocation interfering compounds. US Patent Application No. 20040086904 (2004)
Kurzchalia, T. V. et al. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature 320, 634–636 (1986)
Krieg, U. C., Walter, P. & Johnson, A. E. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl Acad. Sci. USA 83, 8604–8608 (1986)
Voigt, S., Jungnickel, B., Hartmann, E. & Rapoport, T. A. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 134, 25–35 (1996)
Wolin, S. L. & Walter, P. Discrete nascent chain lengths are required for the insertion of presecretory proteins into microsomal membranes. J. Cell Biol. 121, 1211–1219 (1993)
Gorlich, D., Hartmann, E., Prehn, S. & Rapoport, T. A. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature 357, 47–52 (1992)
Fons, R. D., Bogert, B. A. & Hegde, R. S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 160, 529–539 (2003)
Deshaies, R. J., Sanders, S. L., Feldheim, D. A. & Schekman, R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349, 806–808 (1991)
Panzner, S., Dreier, L., Hartmann, E., Kostka, S. & Rapoport, T. A. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81, 561–570 (1995)
Hamman, B. D., Hendershot, L. M. & Johnson, A. E. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell 92, 747–758 (1998)
Lyman, S. K. & Schekman, R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell 88, 85–96 (1997)
Tyedmers, J., Lerner, M., Wiedmann, M., Volkmer, J. & Zimmermann, R. Polypeptide-binding proteins mediate completion of co-translational protein translocation into the mammalian endoplasmic reticulum. EMBO Rep. 4, 505–510 (2003)
Gorlich, D. & Rapoport, T. A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75, 615–630 (1993)
Mothes, W., Jungnickel, B., Brunner, J. & Rapoport, T. A. Signal sequence recognition in cotranslational translocation by protein components of the endoplasmic reticulum membrane. J. Cell Biol. 142, 355–364 (1998)
Plath, K., Mothes, W., Wilkinson, B. M., Stirling, C. J. & Rapoport, T. A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94, 795–807 (1998)
Van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004)
Boger, D. L., Keim, H., Oberhauser, B., Schreiner, E. P. & Foster, C. A. Total synthesis of HUN-7293. J. Am. Chem. Soc. 121, 6197–6205 (1999)
Acknowledgements
E.J.K. thanks E. Rosler, S. Privat, D. Nguyen, J. Melrose, M. Fischer and M. Dea for technical assistance. R.S.H. thanks A. Sharma and M. Alken for technical assistance, and N. Rane and E. Snapp for discussions. J.L.G. thanks Z. Knight for discussions and critical reading of the manuscript. J.L.G. is supported by an NSF graduate fellowship. J.T. is a Searle Scholar and a fellow of the Alfred P. Sloan Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure S1
Effect of cotransin and nor-cotransin on protein expression in stimulated primary human cells. (PDF 19 kb)
Supplementary Figure S2
Effect of cotransin and nor-cotransin on cellular glycoprotein expression. (PDF 1888 kb)
Supplementary Figure S3
Cotransin does not influence the translocation, glycosylation, membrane insertion, or topology of model membrane proteins synthesized in vitro. (PDF 1174 kb)
Supplementary Figure S4
Cotransin's effects are independent of ER lumenal proteins. (PDF 1266 kb)
Supplementary Figure S5
Analysis of VCAM1 and pPrl translocation in fractionated proteoliposomes. (PDF 256 kb)
Supplementary Figure S6
Purification of Sec61 complex / SR and preparation of minimal proteoliposome. (PDF 381 kb)
Supplementary Table 1
Additional full length proteins assayed for translocation in the presence of cotransin in vitro. (PDF 11 kb)
Supplementary Figure Legends
Text to accompany the above Supplementary Figures. (DOC 46 kb)
Supplementary Methods
Methods and References to accompany Supplementary Figures. (DOC 27 kb)
Rights and permissions
About this article
Cite this article
Garrison, J., Kunkel, E., Hegde, R. et al. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature 436, 285–289 (2005). https://doi.org/10.1038/nature03821
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03821
This article is cited by
-
A common mechanism of Sec61 translocon inhibition by small molecules
Nature Chemical Biology (2023)
-
Ipomoeassin-F disrupts multiple aspects of secretory protein biogenesis
Scientific Reports (2021)
-
Substrate Specific Inhibitor Designed against the Immunomodulator GMF-beta Reversed the Experimental Autoimmune Encephalomyelitis
Scientific Reports (2020)
-
Prospects for pharmacological targeting of pseudokinases
Nature Reviews Drug Discovery (2019)
-
Inhibitors of protein translocation across membranes of the secretory pathway: novel antimicrobial and anticancer agents
Cellular and Molecular Life Sciences (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.