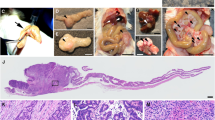
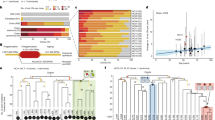
Abstract
Retroviruses, acting as somatic cell insertional mutagens, have been widely used to identify cancer genes in the haematopoietic system and mammary gland1,2. An insertional mutagen for use in other mouse somatic cells would facilitate the identification of genes involved in tumour formation in a wider variety of tissues. Here we report the ability of the Sleeping Beauty transposon to act as a somatic insertional mutagen to identify genes involved in solid tumour formation. A Sleeping Beauty transposon, engineered to elicit loss-of-function or gain-of-function mutations, transposed in all somatic tissues tested and accelerated tumour formation in mice predisposed to cancer. Cloning transposon insertion sites from these tumours revealed the presence of common integration sites, at known and candidate cancer genes, similar to those observed in retroviral mutagenesis screens. Sleeping Beauty is a new tool for unbiased, forward genetic screens for cancer genes in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Neil, J. C. & Cameron, E. R. Retroviral insertion sites and cancer: fountain of all knowledge? Cancer Cell 2, 253–255 (2002)
Jonkers, J. & Berns, A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim. Biophys. Acta 1287, 29–57 (1996)
Ivics, Z., Hackett, P. B., Plasterk, R. H. & Izsvak, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 (1997)
Dupuy, A. J., Fritz, S. & Largaespada, D. A. Transposition and gene disruption in the male germline of the mouse. Genesis 30, 82–88 (2001)
Horie, K. et al. Efficient chromosomal transposition of a Tc1/mariner-like transposon Sleeping Beauty in mice. Proc. Natl Acad. Sci. USA 98, 9191–9196 (2001)
Fischer, S. E., Wienholds, E. & Plasterk, R. H. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl Acad. Sci. USA 98, 6759–6764 (2001)
Yant, S. R. et al. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nature Genet. 25, 35–41 (2000)
Montini, E. et al. In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol. Ther. 6, 759–769 (2002)
Abdallah, B. et al. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Hum. Gene Ther. 7, 1947–1954 (1996)
Hawley, R. G., Lieu, F. H., Fong, A. Z. & Hawley, T. S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1, 136–138 (1994)
Cherry, S. R., Biniszkiewicz, D., van Parijs, L., Baltimore, D. & Jaenisch, R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol. Cell. Biol. 20, 7419–7426 (2000)
Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 (1997)
Kamijo, T. et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91, 649–659 (1997)
Kamijo, T., Bodner, S., van de Kamp, E., Randle, D. H. & Sherr, C. J. Tumor spectrum in ARF-deficient mice. Cancer Res. 59, 2217–2222 (1999)
Wu, X., Li, Y., Crise, B. & Burgess, S. M. Transcription start regions in the human genome are favored targets for MLV integration. Science 300, 1749–1751 (2003)
Vigdal, T. J., Kaufman, C. D., Izsvak, Z., Voytas, D. F. & Ivics, Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 323, 441–452 (2002)
Carlson, C. M. et al. Transposon mutagenesis of the mouse germline. Genetics 165, 243–256 (2003)
Horie, K. et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 23, 9189–9207 (2003)
Mikkers, H. et al. High-throughput retroviral tagging to identify components of specific signalling pathways in cancer. Nature Genet. 32, 153–159 (2002)
Johansson, F. K. et al. Identification of candidate cancer-causing genes in mouse brain tumors by retroviral tagging. Proc. Natl Acad. Sci. USA 101, 11334–11337 (2004)
Dupuy, A. J., Akagi, K., Largaespada, D. A., Copeland, N. G. & Jenkins, N. A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature doi:10.1038/nature03691 (this issue)
Akagi, K., Suzuki, T., Stephens, R. M., Jenkins, N. A. & Copeland, N. G. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 32, D523–D527 (2004)
Lund, A. H. et al. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nature Genet. 32, 160–165 (2002)
Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002)
Seidel, C. et al. Alterations of cancer-related genes in soft tissue sarcomas: hypermethylation of RASSF1A is frequently detected in leiomyosarcoma and associated with poor prognosis in sarcoma. Int. J. Cancer 114, 442–447 (2005)
Heidecker, G. et al. Mutational activation of c-raf-1 and definition of the minimal transforming sequence. Mol. Cell. Biol. 10, 2503–2512 (1990)
Geurts, A. M. et al. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol. Ther. 8, 108–117 (2003)
Acknowledgements
We thank M. Roussel for providing Arf-/- mice; P. Woll, S. Rosenthal, R. Wang, P. Lobitz, J. Zinggeler and M. Davies for technical assistance; P. Hackett, S. McIvor, S. Ekker, P. Marker and members of the Largaespada laboratory for critical reading of the manuscript; N. Kirchhof for performing the pathology; T. L. Bergemann for providing assistance with statistical analysis; and K. Akagi for performing automated sequence analysis and BLAT searches. T2/Onc transgenic mice were generated by the University of Minnesota Mouse Genetics Laboratory. This work was supported by the Arnold and Mabel Beckman Foundation, the National Institute of Drug Abuse and the National Cancer Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
D.A.L. is a founder of Discovery Genomics, Inc. (DGI), a biotechnology company that has licensed SB technology from the University of Minnesota for use in human gene therapy. None of the work reported in this manuscript involved DGI.
Supplementary information
Supplementary Methods
Primer sequences. (DOC 22 kb)
Supplementary Table S1
Details of integrations in CISs. (DOC 366 kb)
Supplementary Table S2
Several T2/onc integrations occurred near previously identified leukaemia retroviral CISs. (DOC 20 kb)
Supplementary Figure S1
T2/onc Southern analysis. (PDF 143 kb)
Supplementary Figure S2
T2/onc integrations are tumour specific. (PDF 68 kb)
Rights and permissions
About this article
Cite this article
Collier, L., Carlson, C., Ravimohan, S. et al. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436, 272–276 (2005). https://doi.org/10.1038/nature03681
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03681
This article is cited by
-
CRISPR and transposon in vivo screens for cancer drivers and therapeutic targets
Genome Biology (2020)
-
A simplified transposon mutagenesis method to perform phenotypic forward genetic screens in cultured cells
BMC Genomics (2019)
-
How to Choose a Mouse Model of Breast Cancer, a Genomic Perspective
Journal of Mammary Gland Biology and Neoplasia (2019)
-
PiggyBac transposon tools for recessive screening identify B-cell lymphoma drivers in mice
Nature Communications (2019)
-
Loss-of-function genetic tools for animal models: cross-species and cross-platform differences
Nature Reviews Genetics (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.