Abstract
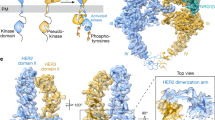
HER2 (also known as Neu, ErbB2) is a member of the epidermal growth factor receptor (EGFR; also known as ErbB) family of receptor tyrosine kinases, which in humans includes HER1 (EGFR, ERBB1), HER2, HER3 (ERBB3) and HER4 (ERBB4)1. ErbB receptors are essential mediators of cell proliferation and differentiation in the developing embryo and in adult tissues2, and their inappropriate activation is associated with the development and severity of many cancers3. Overexpression of HER2 is found in 20–30% of human breast cancers, and correlates with more aggressive tumours and a poorer prognosis4. Anticancer therapies targeting ErbB receptors have shown promise, and a monoclonal antibody against HER2, Herceptin (also known as trastuzumab), is currently in use as a treatment for breast cancer5. Here we report crystal structures of the entire extracellular regions of rat HER2 at 2.4 Å and human HER2 complexed with the Herceptin antigen-binding fragment (Fab) at 2.5 Å. These structures reveal a fixed conformation for HER2 that resembles a ligand-activated state, and show HER2 poised to interact with other ErbB receptors in the absence of direct ligand binding. Herceptin binds to the juxtamembrane region of HER2, identifying this site as a target for anticancer therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Yarden, Y. & Sliwkowski, M. X. Untangling the ErbB signalling network. Nature Rev. Mol. Cell Biol. 2, 127–137 (2001)
Olayioye, M. A., Neve, R. M., Lane, H. A. & Hynes, N. E. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 19, 3159–3167 (2000)
Tang, C. K. & Lippman, M. E. in Hormones and Signaling (ed. O'Malley, B. W.) 113–165 (Academic, San Diego, 1998)
Slamon, D. J. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 (1987)
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpress HER2. N. Engl. J. Med. 344, 783–792 (2001)
Carpenter, G. Receptors for epidermal growth factor and other polypeptide mitogens. Annu. Rev. Biochem. 56, 881–914 (1987)
Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 103, 211–225 (2000)
Jones, J. T., Akita, R. W. & Sliwkowski, M. X. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 447, 227–231 (1999)
Di Fiore, P. P. et al. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science 237, 178–182 (1987)
Ogiso, H. et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110, 775–787 (2002)
Garrett, T. P. J. et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell 110, 763–773 (2002)
Ferguson, K. M. et al. EGF activates its receptor by relieving auto-inhibition of ectodomain dimerization. Mol. Cell (in the press)
Cho, H. S. & Leahy, D. J. Structure of the extracellular region of HER3 reveals an interdomain tether. Science 297, 1330–1333 (2002)
Banfield, M. J., King, D. J., Mountain, A. & Brady, R. L. VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs. Proteins 29, 161–171 (1997)
Lawrence, M. C. & Colman, P. M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 (1993)
Berezov, A. et al. Disabling receptor ensembles with rationally designed interface peptidomimetics. J. Biol. Chem. 277, 28330–28339 (2002)
Harari, D. & Yarden, Y. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene 19, 6102–6114 (2000)
Sliwkowski, M. X. et al. Nonclinical studies addressing the mechanism of action of Trastuzumab (Herceptin). Semin. Oncol. 26, 60–70 (1999)
Molina, M. A. et al. Trastuzumab (Herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res. 61, 4744–4749 (2001)
Burke, C. L., Lemmon, M. A., Coren, B. A., Engelman, D. M. & Stern, D. F. Dimerization of the p185neu transmembrane domain is necessary but not sufficient for transformation. Oncogene 14, 687–696 (1997)
Denney, D. W. Jr Gene amplification methods. US patent 5,776,746 (1998).
Leahy, D. J., Dann, C. E., Longo, P., Perman, B. & Ramyar, K. X. A mammalian expression vector for expression and purification of secreted proteins for structural studies. Protein Expr. Purif. 20, 500–506 (2000)
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997)
Jones, T., Zou, J.-Y., Cowan, S. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Winn, M. D., Isupov, M. N. & Murshudov, G. N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D 57, 122–133 (2001)
Laskowski, R. A. et al. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
Carson, M. Ribbons. Methods Enzymol. 277, 493–505 (1997)
Kraulis, P. J. A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Acknowledgements
We thank C. Ogata and M. Becker for assistance at beamlines X4A and X25, respectively, of NSLS at Brookhaven National Laboratory; A. Ullrich for supplying a human HER2 complementary DNA; P. Longo for technical assistance; T. Garrett, S. Yokoyama and colleagues for supplying preprints in advance of publication; S. Yokoyama for coordinates of the EGF–EGFR complex; M. Lemmon, K. Ferguson, M. Amzel, J. Berg, S. Bouyain and W. Yang for discussion and comments on the manuscript; A. Guarne for help with figures; and N. Davidson for assistance with Herceptin. This work was supported by the NIH and the HHMI.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Cho, HS., Mason, K., Ramyar, K. et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 421, 756–760 (2003). https://doi.org/10.1038/nature01392
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01392
This article is cited by
-
Effects of Chemical Fixatives on Kinetic Measurements of Biomolecular Interaction on Cell Membrane
The Journal of Membrane Biology (2024)
-
A bispecific antibody targeting HER2 and CLDN18.2 eliminates gastric cancer cells expressing dual antigens by enhancing the immune effector function
Investigational New Drugs (2024)
-
Preclinical development of a first-in-class vaccine encoding HER2, Brachyury and CD40L for antibody enhanced tumor eradication
Scientific Reports (2023)
-
Label-free electrochemical cancer cell detection leveraging hemoglobin-encapsulated silver nanoclusters and Cu-MOF nanohybrids on a graphene-assisted dual-modal probe
Scientific Reports (2023)
-
Biased activation of the receptor tyrosine kinase HER2
Cellular and Molecular Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.