Abstract
Major programs in psychiatric genetics have identified >150 risk loci for psychiatric disorders. These loci converge on a small number of functional pathways, which span conventional diagnostic criteria, suggesting a partly common biology underlying schizophrenia, autism and other psychiatric disorders. Nevertheless, the cellular phenotypes that capture the fundamental features of psychiatric disorders have not yet been determined. Recent advances in genetics and stem cell biology offer new prospects for cell-based modeling of psychiatric disorders. The advent of cell reprogramming and induced pluripotent stem cells (iPSC) provides an opportunity to translate genetic findings into patient-specific in vitro models. iPSC technology is less than a decade old but holds great promise for bridging the gaps between patients, genetics and biology. Despite many obvious advantages, iPSC studies still present multiple challenges. In this expert review, we critically review the challenges for modeling of psychiatric disorders, potential solutions and how iPSC technology can be used to develop an analytical framework for the evaluation and therapeutic manipulation of fundamental disease processes.
Similar content being viewed by others
A need for disease models
Psychiatric disorders are associated with major economic, societal and personal burdens. As a group, they constitute 13% of the global burden of disease, and are the leading cause of disability worldwide.1, 2 Multiple lines of investigation from brain imaging, studies of post-mortem brain tissue and genetic studies implicate aberrant cellular function in the most serious psychiatric disorders (for example, schizophrenia (SCZ), bipolar disorder, autism spectrum disorder (ASD), anorexia nervosa and major depressive disorder). However, these implications have not been tested in vitro, and this relative lack of understanding of disease mechanisms hampers the development of treatment. Induced pluripotent stem cells (iPSC) technology is an exciting and very promising tool to generate new disease models, with the ultimate goal of creating a new generation of pathophysiology-relevant assays for in vitro drug screening.3 iPSC-based investigation has added advantages of permitting temporal analyses of neurodevelopmental deficits that are not as readily available in animal studies and human studies, allowing longitudinal cell studies that follow the progress of disease processes from initiation to their end point.4, 5, 6, 7, 8 However, to develop iPSC-based assays that truly reflect the pathophysiology of psychiatric disorders, we need a precise understanding of which molecular pathways and cellular structures are involved.
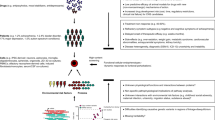
Here we review the search for cellular models and phenotypes in the context of the current state of the art for SCZ genetics and understanding gained from SCZ-related animal models (Figure 1). We will discuss current capabilities and further developments needed, potential pitfalls for stem cells reprogramming, culturing and in vitro differentiation; and the establishment of relevant cellular phenotypes that can be translated into disease models (Figure 1), and ultimately into pharmaceutical targets for psychiatric disorders.
Brain correlates of in vitro iPSC cell phenotypes. Current analysis of patient iPSC offers a range of potential methods of cell phenotyping that correlates to potential changes in brain pathology associated with psychiatric disorders. Gene expression (transcriptomic) and protein expression (proteomic) profiling of in vitro neurodevelopment or iPSC-derived mature neuronal and glial cultures correlates with brain development and processes associated with adult neurogenesis, such as some aspects of memory and learning. Cell analysis of neurodevelopment, cell migration, cytoskeletal dynamic and synaptogenesis informs on the basic processes by which neurogenesis builds and remodels the brain. Functional activity is measured by electrophysiological recording (for example, patch-clamp) and calcium or voltage sensors (dyes and genetically encoded markers). Multicellular interactions (connectomics) can be investigated as structural interactions in two-dimensional (2D), 3D and organoid cell co-culture, and at the functional level using multi-electrode and microelectrode array (MEA) recordings. iPSC, induced pluripotent stem cell.
Advances in psychiatric genomics: the case of SCZ
Decades of twin/family studies have compellingly established that psychiatric disorders are heritable.9 However, the identification of causal genetic variants has, until recently, been notably difficult. Unprecedented advances in the past decade have shown that psychiatric disorders are complex and influenced by the combination of hundreds of common genetic variants each of relatively small impact on disease risk and occassionally by rare variants with larger effects.10 The field has made major advances in identification of these risk variants, although it is clear that there are more to be found, and how they combine together to create a polygenic risk is currently unknown.
Genetic epidemiology provides strong support for a genetic component for SCZ (with a heritability of ~0.64 in Nordic population samples and 0.81 in a twin study meta-analysis).9, 10, 11, 12, 13 Common variation assessed by genome-wide association studies (GWAS) yields single nucleotide polymorphism-based heritability estimates for SCZ over 0.30, and have yielded 108 independent genomic risk loci.14, 15 Critically, most loci identified in GWAS are broad (median 129 kb) with small impact on SCZ risk (median relative risk 1.08).14 Whole-exome sequencing studies for SCZ identified no specific genes but implied a role for functional gene sets, for example, voltage-gated calcium channels, ARC-associated scaffold and FMRP interactors.
Copy number variation (CNV) studies for SCZ have yielded a dozen CNVs that are robustly associated with SCZ, but also with other psychiatric disorders.10, 16, 17 Findings from recent studies suggest that a high polygenic burden adds to the SCZ risk in carriers of CNVs, suggesting cumulative effects between common and rare risk variants.14, 18, 19, 20
SCZ, but also ASD, converge on common pathways; such as within synaptogenesis and synapse function21, 22 and epigenetic processes,22, 23 with many genes being highly expressed during fetal cortical development.24, 25, 26
Despite these unprecedented advances in the genetics of SCZ, very few of the current findings unequivocally implicate specific individual genes that are easily ‘actionable’ for biological, clinical or therapeutic studies. To be of value, such studies need to show strong linkage between the genetic variation and a discriminative phenotype that is relevant for the disorder. This connection is both crucial for understanding the molecular pathways that lead to SCZ and essential to develop iPSC-based assays that reflect SCZ pathophysiology. A single variant of small effect is unlikely to yield a measurable cellular phenotype, we therefore aim to model either the cumulative effect of hundreds of risk variants of small effect or a single variant of high penetrance or large effect.
iPSC technology27, 28 has provided a highly promising tool to investigate human disorders, and is especially well-suited to deal with disorders that are not caused by a single mutation, such as mental disorders.4, 29 As iPSC studies rely on cells from patients, one can select patients with a high genetic propensity for the disease, either due to the accumulation of many common variants of small effect or due to carrier status of a rare variant of large effect. In addition, by utilizing cells from patients with a targeted set of risk alleles, one also captures the complete genetic background of an individual, which includes possible genetic modifiers that are currently unidentified.4 Several initial iPSC studies for SCZ have already been carried out and have yielded proof-of-principle by successfully identifying differences in synaptic functions in iPSC-derived cells from patients. However, these initial studies also clearly illustrate some of the pitfalls of iPSC studies for identifying cellular traits associated with SCZ. We will discuss these pitfalls in more detail below, after first examining the alternative approach of using animal models for SCZ and ASD.
Rodent models in the study of SCZ and autism
Several approaches have been employed to produce rodent models for SCZ and ASD. Rodents provide a number of general advantages with respect to animal husbandry and handling, well-established behavioral and physiological tests, and the availability of transgenic manipulation.30, 31 For example, maternal stress and malnutrition, infection and hypoxic insult at birth have all been implicated as developmental triggers of SCZ, and these can be replicated in rodent models through manipulations such as prenatal drug administration, disruption of neurogenesis during gestational periods, neonatal ventral hippocampal lesions, post-weaning social isolation and perinatal or maternal immune activation (reviewed in ref. 30). Genetic manipulation has also been employed to target several genes implicated in SCZ or ASD in transgenic mice.31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42 An important caveat with respect to transgenic models is that these are relatively easy to generate, and thus may fuel research efforts that are actually red herrings because the genes in question are not convincingly implicated in the human diseases.43
Although rodents provide tractable and accessible platforms, these are not without significant shortcomings. First, although each of the rodent models that are already established replicates certain neurophysiological, neuroanatomical and/or behavioral features of genetic mutations implicated in SCZ or ASD, none of them fully recapitulates the complexity of these disorders. Thus, the knowledge contributed by animal models to the etiology of SCZ and ASD is by its nature fragmentary, with each model providing a specific facet that needs to be integrated into a greater whole that reflects the heterogeneity implicit in the disorder itself. Second, animal models cannot be interrogated with the necessary depth. Indeed, how can one gauge effects on thought processes, perception and abstract learning in animals, and when these can only be conveyed fully through language? As a consequence, many core features of psychiatric disorders can only be assessed indirectly or obliquely in animal models, with an artificial focus on simpler behavioral and physiological features that can be easily identified. The extent to which these can be translated to the more complex symptomatology of the human conditions is not always clear. Third, induction of disease states in rodents may involve acute pharmacological or other insults that do not accurately replicate the causes of psychiatric disorders in humans. Even transgenic approaches targeting the same genes may be inaccurate, since a manipulation as coarse as a single gene knockout is unlikely to capture the complexity of the genetic causes of psychiatric disorders. Moreover, the genomic landscapes of the genes in question may differ in rodents and humans, and genetic differences will also be compounded where genes and environment interact extensively in disease development. Fourth, rodents and humans have vastly different lifespans, which may not be appropriately congruent with respect to the timeline of disease development. Last, the pharmacology of potential drug treatments may differ in the two species, creating false positives and negatives in preclinical studies.
In conclusion, only a small percentage of psychiatric disorders is caused by single gene variations and can be modeled with transgenic mice. Indeed, many clinical studies based on promising drug targets found in animals failed human translation. As a consequence, the difficulties in modeling polygenic risk gene variants and the human genetic background have made animal models less attractive in modeling complex neurological disorders. Nevertheless, transgenic mouse models advanced our understanding of potential mechanisms regulated by genes involved in psychiatric disorders. Table 1 compares some of the principal advantages of transgenic mouse models and human iPSC-based models.
Cell phenotyping of patient iPSC
The key challenge for iPSC-based disease modeling is to identify one or more relevant cellular phenotypes that accurately represent the disease pathophysiology. Increasing numbers of reports have demonstrated that for many diseases specific pathophysiology can be captured in human iPSC-based disease models. These range from cardiovascular disease,44, 45 cancer,46, 47 ocular disease,48, 49 diabetes mellitus50, 51 and neurological disorders of the brain.52, 53 Can the same approach be applied to complex psychiatric disorders?
The problem is that almost all psychiatric disorders are characterized by clinical signs and symptoms, but lack independent verification from objective biomarkers. Thus, how might these clinical phenotypes manifest themselves in terms of cell behavior? The identity of robust cellular ‘readouts’, which typify any psychiatric disorder, is a crucial unsolved problem and an area of intense study54 (Table 2). When satisfactorily answered, this will herald a new degree of biological objectivity and quantification for the study of psychiatric disorders.
The aim is to find a single or small number of cell phenotypes or parameters that strongly associate with psychiatric disorders, and establish a cellular profile characteristic of cells derived from the general patient population. Although a consensus set of cellular phenotypes for psychiatric disorder is yet to be established, we can define some of their desired characteristics. First, cellular phenotypes have to relate to the biological pathways identified by genetics. Second, although there are many risk genes in disparate biological pathways, at some level, phenotypes should converge onto a much smaller grouping. Third, phenotypes need to be quantifiable. Finally, to be useful for drug development cellular phenotypes should be reversed by pharmacological treatment, although not necessarily by drugs in current use.
Although human iPSC-based approaches underrepresent the complexity of the human central nervous system, cellular phenotypes are likely to lie more proximal to molecular disease mechanisms than phenotypes seen at the level of a tissue or organism,55 and thus may bypass compensatory homeostatic processes that buffer the effects of deleterious genetic variants in whole tissues and organs (Table 2). Identification of cellular phenotypes may therefore offer a more direct readout of the pathophysiological process (Figure 1). This, of course, would need to be validated against clinical data. We discuss below various ways to phenotype iPSC-derived patients cells and to interrogate the phenotypes to extract information about the disorder.
‘Omic’ approaches to cell phenotyping
Post-genomic technologies offer a battery of approaches for profiling cell difference at both population and single-cell level. Advances in RNA sequencing technologies and transcriptomics provide one of the easiest and highest throughput approaches to cell phenotyping, and potentially could be compared with transcription profiles from brain biopsies or post-mortem tissue. Transcriptome studies of both SCZ and ASD patient-derived cells have identified hundreds of gene expression differences.56, 57, 58, 59 To date, however, there is no definitive or consensus RNA-based transcription profile associated with SCZ or ASD-derived iPSC, a situation that is mirrored by post-mortem brain tissue profiling.60 Trends point to expression changes of genes involved in synaptic structure, adhesion and transmission, and specific cell signaling pathways particularly those associated with glutamate, Wnt and cAMP signaling.61 These studies also often reveal overlap with risk genes identified by both GWAS and CNV studies, and may point to convergence on biological pathways rather than on individual genes.
Mapping and measuring DNA methylation may extend this analysis to provide unique epigenetic signatures. For example, the methyl-cytosine-binding protein MeCP2 is causative of Rett’s syndrome and is associated with ASD.62 Histone protein modifications can be profiled using ChIP-seq and a number of histone methyl transferase enzymes are associated with neuropsychiatric disorders. Either alone, or more likely when combined with expression data, epigenetic profiling may identify developmental and activity-dependent cellular phenotypes.63, 64, 65
Proteomic technologies have also been used to investigate patient iPSC phenotypes, backing up the results of transcriptional profiling. Use of SILAC (stable isotope labeling by amino acids in cell culture) in mass spectrometry to quantitate changes in protein levels of neural progenitor cells (NPC) has shown decreases in the SCZ-associated NLGN3 protein, and increases in the actin cytoskeletal regulators Cofilin and Profilin, and proteins associated with oxidative stress.66 Protein profiling can be extended further to protein function by investigating protein interactomes and phosphoproteomes. An analysis of the protein complexes in neuron-like SH-SY5Y cells showed a convergence of proteins encoded by ASD associated genes onto a small number of protein complexes.67 Phosphoproteomic analysis of iPSC-derived from Phelan–McDermid syndrome (PMDS) patients, in which the post-synaptic density protein SHANK3 is lost, and of neurons from Shank3 knockdown mice, has revealed elevated activity of protein kinase CLK2 and demonstrated its potential as a therapeutic target.68
Neurodevelopmental deficits
A major advantage of patient-derived iPSC studies is the possibility to follow neurodevelopment in vitro. Transcriptional analyses are beginning to show that differences in neurodevelopment may arise prior to overt neuronal differentiation, leading to altered timing or cell differentiation fates of NPC.66 This fits with the potential neurodevelopmental component of psychiatric conditions and the profile of genetic risk. Transcriptional differences can readily be confirmed using the extensive range of antibodies to neurodevelopmental marker proteins. Interestingly, a number of CNV cases, such as PMDS and Timothy Syndrome, in which there is a deficit in the voltage-gated calcium channel, CACNA1C 57 show gene expression changes at the NPC stage. This may reflect a feedback onto neurodevelopmental signaling due to electrophysiological activity or cell contact-dependent gene expression69 in determining developmental timing and differentiation. Such studies raise the possibility that transcriptional profiling of developmentally regulated gene expression in patient-derived iPSC may reveal quantitatively robust and disease-relevant phenotypes.
In addition to neuronal deficits, abnormalities of all three glial cell types have been observed in SCZ patients.70 Post-mortem studies indicate that oligodendrocyte numbers are reduced, and that oligodendrocyte maturation and morphology is impaired in SCZ patients,71, 72 although neuroleptic treatment and aging might have confounded these results. Results from large-scale expression analyses and GWAS for SCZ,73, 74, 75 implicate changes in genes that regulate cell-cycle control and oligodendrocyte maturation, suggesting impaired cell-cycle exit and re-entry.71 Altered astrocyte numbers are also found in the brains of SCZ patients after autopsy with early studies reporting astrogliosis,76 while more recent studies indicate astrocyte cell loss in selected (sub)cortical and callosal regions.77 GWAS demonstrates genetic variants in genes involved in astrocyte function, including signal transduction, tyrosine kinase signaling, G protein–coupled receptor signaling, small GTPase-mediated signaling, cell adhesion and gene transcription.73 These findings are supported by results from expression studies that showed altered expression levels of astrocyte-associated genes, including GFAP,78 glutamine synthetase79 and S100B.80 The involvement of biological pathways associated with inflammation and immunity in the development of SCZ is receiving increased attention, and is supported by patient genetic studies, with variants found in several cytokine genes,81, 82 as well as the major histocompatibility complex region83 where structural variants of the complement component 4 (C4) gene lead to increased activity.84 Previous studies have indicated microglial activation and altered microglia-related gene expression in postmortem brain tissue (reviewed by ref. 70). As severe infections and aberrant immune responses are risk factors for SCZ, this may point to gene–environment interactions for SCZ and the use of anti-inflammatory drugs in treatment strategies.85
Altered neuronal cell biology and function
Beyond transcriptomics and neurodevelopment, other cell parameters may also be effective measures of cellular phenotype. Brennand et al.66 noted reduced migration in neural precursor cells from four SCZ patients, using a variety of assay formats.66 The molecular mechanism for this is not entirely clear but correlates with increased expression of adhesion molecules. Its relevance may be significant, since interneurons migrate from specific progenitor domains to populate cortical and other regions during brain development. Changes in morphology due to alterations of the cytoskeleton have also been observed, suggesting more than just an adhesion effect.86 Cytoskeletal effects could manifest later in development as changes in neuroarchitecture and particularly in dendritic spine morphology and dynamics. If of sufficient magnitude, such structural sequelae could lead to macroscopic changes in brain anatomy that may correlate with larger scale changes detected by human brain imaging.87 Computed tomography and magnetic resonance imaging analyses have revealed structural changes in the brain of SCZ patients, such as enlarged lateral and third ventricles, smaller cortical volumes, smaller gray matter volumes and larger basal ganglia (reviewed by ref. 88). Morphometric changes generally do not progress overtime and therefore may match histological findings in postmortem tissue, which include altered cortical and hippocampal pyramidal neuron size, decreased interneuron numbers and reduced dendritic spine densities.88, 89
At the subcellular level, altered synaptogenesis, synaptic vesicle release and mitochondrial function have all been observed in patient-derived iPSCs.58, 62 Recently, mitochondrial abnormalities have been detected using RNA sequencing and mitochondrial assays, and hyperexcitability has been demonstrated by using both patch-clamp recording and Ca2+ imaging in immature neurons from patients with bipolar disorder.90
The gold standard for electrophysiological assessment is patch clamping recording, and mature differentiated neurons are assessable with this technique. However, this approach requires obtaining high-resistance seals between the electrode tip and the neuron surface for full effect, limiting throughput, even in automated systems. An alternative is to use optical recording of electrical events to monitor cell activity. This is most commonly done indirectly by imaging calcium fluctuations, using calcium sensitive fluorescent dyes or genetically encoded calcium indicators.91 The latter approach has the added advantage that genetically encoded calcium indicators can be selectively expressed in specific cell types. Calcium recording, however, can only capture events that involve changes in intracellular calcium concentration, meaning that hyperpolarization and inhibitory synaptic events go largely unrecognized, and it has low-temporal resolution. Voltage-sensitive indicators on the other hand provide direct information about changes in membrane potential irrespective of the cause and the sign, permitting the assessment of both excitatory and inhibitory synaptic interactions and depolarizing and hyperpolarizing neurotransmitter and drug effects.92, 93
Currently, the range of phenotypes seen in cellular studies of neuropsychiatric disorders is diverse and variable, and there is a need for a more systematic investigation across a range of phenotypes. For example, effects on single-cell electrophysiological parameters appear variable with no clear pattern. What is clear, however, is that there do not seem to be major deficits in the basic electrophysiological behavior of neurons. Where differences are emerging is in synaptic function and connectivity, findings that fit the types of pathways implicated by genetic analysis. This may point to the major problem with single-cell measurements, as the biology of neuropsychiatric disorders is ultimately an emergent property of cell connectivity and network activity.
Cell interactions and neural networks
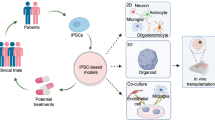
Two general approaches can be taken to assay neural networks and cell connectivity; one focuses on structural interaction, the other on functional connectivity, although ideally these could be combined to provide both structural and functional assemblies. Conventional two-dimensional (2D) monocultures are limited in their ability to form dynamic anatomical connections and may not follow the same neurodevelopmental pathway, as cells within the brain are both constrained by, and receiving signals from the extracellular matrix and neighboring cells. 3D culture methods are being explored to better mimic tissue architecture, and to study cellular properties and network interactions in health and disease. Ideally, these iPSC-based 3D culture platforms would involve co-cultures of appropriate neuronal and glial cell types in a mechanically appropriate matrix with soluble and extracellular matrix-derived signals to those extant in the developing brain. They would also be compatible with optical imaging for morphological and electrophysiological analysis. Current approaches are based on the use of biomaterials to support 3D network organization and/or the use of neural stem cell aggregates to reconstruct complex in vivo-like structures (organoids). The range of biocompatible materials being tested for 3D culture systems includes hydrogel-based materials, 3D electrospun polymers, synthetic scaffolds, silica beads and microfluidic bioreactors. At present, we lack a full understanding of how biomaterials affect cell properties, and 3D cultures systems have so far not been widely explored for iPSC-based disease modeling.94, 95, 96 Organoids and aggregate cultures give prospects to reconstruct in vivo-like neural circuits and to achieve insights into the signal integrated on multiple levels.97 Through self-organization of complex tissue patterns, attempts have been made to replicate various brain regions to generate models of ASD.98, 99, 100 Of particular note, 3D human ‘cortical spheroids’ generate a laminated cerebral cortex-like structure containing electrophysiologically mature neurons that form functional synapses.98 However, it is difficult to control the size and internal laminar structure of the spheroids, and robust and reproducible methods need to be developed for quantification of both structure and physiology.
Functional network studies have substantial potential as drug-screening platforms. Ideally, they should measure such network behavior as the degree of connectivity (spread of impulses through the network) and the synchrony and oscillation frequency of neuronal firing.101 In principle, these could mirror at the cellular level the types of brain activity measured by electroencephalogram, but at much higher spatial resolution. Human iPSCs can be developed into functional neuronal network on in vitro microelectrode arrays,102, 103, 104 where network behavior has been shown to be sensitive to reduced expression of post-synaptic genes associated with SCZ and bipolar disorder.105 Network level properties may be utilized to model the beta and gamma oscillation perturbations observed in patients.106 Calcium- and voltage-sensitive imaging either instead of or in conjunction with microelectrode array recordings provide powerful options to observe activity oscillations in neuronal networks. Many of molecular phenotyping methods, such as transcriptomic and cell morphology analyses can be combined with these electrophysiological assays in high throughput to facilitate a multimodal assessment of many patient-derived iPSC lines.
Challenges for iPSC-based disease models
Modeling psychiatric disorders at the cellular level is not without difficulties and there are many potential sources of error. First, to faithfully model diseases it is important that variation detected between the iPSC lines reflects the underlying genetic differences associated with the disorder, and is not introduced by cell reprogramming or downstream effects of cell culturing and differentiation protocols. This is particularly crucial when cellular differences between cases and controls are expected to be subtle. Substantial experimental variation between iPSC lines can arise from inconsistency in iPSC reprogramming protocols, parental somatic cell type107 and persistent epigenetic modifications;108, 109 interline variability due to genetic instability, mosaicism or accumulation of mutations during cell line expansion;110 and intra-line variability arising during prolonged cell culturing and differences in growth conditions.107, 111, 112, 113 However, methods for reprogramming have steadily improved since Takahashi and Yamanaka27 first described iPSC, and recent studies show that by following standardized protocols reprogramming and cell culturing consistency can be achieved and intra-line variation reduced.114, 115, 116 Evidence for the equivalence of human iPSC and embryonic stem cells indicate that reprogramming can instate a pluripotent state similar to that of the inner cell mass of an early human embryo.117 Interestingly, the epigenetic erasure that occurs during the reprogramming process appears to make gene expression in iPSC more dependent on the genotype compared with gene expression in the cells used for reprogramming, in which interline variability is much larger.118 The maturity of iPSC-derived neurons is a concern in studies of neuronal networks since they initially appear to most closely resemble fetal brain cells.66, 98, 119, 120 Different methods have been developed to promote the maturation of iPSC-derived neurons, that is, transplantation into rodent brains may mature them into GABAergic interneuron for up to 7 months.120 Aging of iPSC-derived dopaminergic neurons by induction of progerin expression revealed disease phenotypes such as pronounced dendrite degeneration, progressive loss of tyrosine hydroxylase expression, enlarged mitochondria and Lewy body-precursor inclusions.121
Second is the question of choosing the best patient and control cell combinations. Early iPSC studies did not always fully consider possible confounders, such as differences in genetic background, unmatched age, sex, and ancestry between patients and control individuals, as well as differences in passage number of the iPSC lines.74 It has become clear that these factors need to be carefully considered when selecting appropriate healthy control iPSC.75 For many studies, samples derived from healthy family members of the patient with similar genetic background but not diagnosed with disease currently represent the most feasible control.77
An ideal control would be an isogenic iPSC line generated by correcting the genetic lesion(s) of the patient-derived iPSC line. In recent years, novel techniques for genome-editing have greatly increased level of efficiency of gene targeting in vitro. Using engineered endonucleases such as zinc finger nucleases, TALENs or CRISPR/Cas9, it is now possible to genome edit iPSC with high specificity.76 For monogenic diseases, isogenic gene-corrected iPSC lines represent an ideal control population. However, for complex disorders with multiple genetic loci contributing to the disease, editing approaches face limitations. Although editing of multiple loci is feasible,122 gene correction of a larger number of disease-associated variants in a single iPSC line remains problematic. In principle, a disease versus control scenario could also be generated by active introduction of candidate mutations into ‘healthy’ iPSC via gene editing. However, at present this route too appears only feasible for diseases with relatively small numbers of highly penetrant mutations. Furthermore, it comes with the significant disadvantage that phenotypic alterations in such in vitro-mutated iPSC cannot be correlated with the clinical history of an individual patient.78
Currently, parallel studies on isogenic gene-corrected iPSC for selected variants and cells derived from unaffected family members will remain the most feasible controls for comparative phenotypic analysis of patient-derived iPSC. These studies could be supplemented by the reverse experiment of using genome editing to introduce additional gene mutations into patient or non-patient cell lines with a high polygenic risk score derived by classic reprogramming to create artificial ‘hyperphenotypes’, where the effects of different patient backgrounds can be studied on highly penetrant disease-associated variants (Figure 2a). However, such approaches still face several limitations such as difficulties in engineering large chromosomal deletions. Further challenges include the large number of single nucleotide polymorphisms in linkage disequilibrium and limited information to guide the choice of relevant variants (Table 3).
Automated production and differentiation of iPSCs. (a) Conventional disease modeling or drug evaluation approaches mostly rely on a small number of disease-specific, as well as control iPSC lines and largely ignore the impact of genetic variability on pathological pathways or drug targets. Parallelization of reprogramming and subsequent differentiation would allow assessing phenotypic variation or to validate candidate drugs on multiple genomic backgrounds, for example, stratified patient or control cohorts. (b) Fully integrated robotic systems such as the StemCellFactory (www.stemcellfactory.de) are expected to allow high-throughput reprogramming and differentiation under controlled and standardized conditions, and thus to minimize line-to-line heterogeneity induced by non-standardized manual handling steps. Kindly provided by Andreas Elanzew, Simone Haupt (Life & Brain, Bonn, Germany) and the Fraunhofer Institute for Production Technology (IPT). iPSC, induced pluripotent stem cell.
Finally, inter-individual variability of patients with similar diagnosis and subtle differences in the clinical disease progression will result in quantitative, and perhaps qualitative, differences in cell phenotypes between iPSC lines derived from different patients.110 It is therefore necessary to have the capacity to handle large-sample sizes for modeling these complex disorders. Comprehensive exploration of the steadily increasing number of risk loci in iPSC-based models will only be possible using large cohorts of patients and controls. To assess the combined impact of genetic variants on a single background or to decipher the single contribution of each variant, it will be necessary to explore novel technical solutions that enable much higher throughput. To that end, automated modules covering key reprogramming steps such as transfection, media changes, splitting and colony picking are already being implemented.123 It is foreseeable that automation will move towards large-system integrations enabling fully automated production of iPSC on industrial production-line platforms such as the StemCellFactory (www.stemcellfactory.de; Figure 2b). While automated cell culture provides key advantages with respect to standardization and parallelization, large-system integration units for robotic reprogramming come with their own challenges (Table 4).
Conclusions and future perspectives
The proof-of-concept emerging from many recent studies that have attempted to mimic aspects of psychiatric disorders in vitro using patient-derived cells is very encouraging (Table 2). Increased standardization, proper controls and new integrative robotic systems will give solutions to many problems. However, there remain a number of considerable challenges ahead.
Strategies moving forward need to take into account the genetic characteristics of the patient population in which genetic risk is largely polygenic, and a mixture of many common variants of small effect, as well as few rare variants of large effect. In contrast, a priori we would expect to find the most robust phenotypes in cells derived from patients carrying highly genetically penetrant rare variants and cell models created using genome editing of isogenic iPSC lines. It will be important to connect the knowledge gained from single gene deficits and that gained from the accumulated effects of multiple subtle genetic risk alleles.
Both the selection of patients carrying rare variants of large effect and the selection of patients of extremely high polygenic risk require large patient populations to optimize the selection. When genetic risk in selected patients is not sufficiently causal, any iPSC experiment will require the analysis of large numbers of patient cell lines. An important step is to have robust protocols for reprogramming and differentiation of large numbers of patient samples. This will require standardization and rigorous quality control to reduce technical variation to an acceptable minimum. Given the high current reagent costs for stem cell research, the unit price per patient cell assay needs to drop substantially before this will be feasible. These processes need to integrate well with global efforts in patient recruitment and accompanying clinical phenotyping and genomic analysis.
Beyond the issues of variability and capacity lies the key question of what is the relevant cellular phenotype or phenotypes. We have discussed what is currently possible and under development, and how these might relate to function and physiology in the intact brain. However, these investigations have only just begun, and are likely to require multiple lines of converging evidence, carried out in numerous centers and with validation against clinical and animal model studies, before consensus cellular phenotypes can be established and accepted.
Finally, we need to consider what constitutes success in this enterprise? Although iPSC-based systems provide a powerful route to identifying molecular mechanisms underlying genetic and other disease-related risks, in isolation they do not provide information about brain physiology, higher order neuronal circuitry and function or human psychology. Success might simply be to create a reliable experimental link between genetics and patient studies via cell physiology. Alternatively, we could set more ambitious goals using iPSC to inform connectomic and neuro-computational modeling, predict patient drug responses and promote preclinical drug discovery. True success would be achieved if analysis of iPSC-derived neuronal networks became a standard assay for neurophysiologists, forming an integral component of diagnostic and precision medicine for neuropsychiatric disorders and facilitating the first advent of new drugs screened on patient iPSC reach the clinic.
References
Eaton WW, Martins SS, Nestadt G, Bienvenu OJ, Clarke D, Alexandre P . The burden of mental disorders. Epidemiol Rev 2008; 30: 1–14.
Buka SL . Psychiatric epidemiology: reducing the global burden of mental illness. Am J Epidemiol 2008; 168: 977–979.
Nishikawa S, Goldstein RA, Nierras CR . The promise of human induced pluripotent stem cells for research and therapy. Nat Rev Mol Cell Biol 2008; 9: 725–729.
Marchetto MC, Brennand KJ, Boyer LF, Gage FH . Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum Mol Genet 2011; 20: R109–R115.
Brennand KJ, Simone A, Tran N, Gage FH . Modeling psychiatric disorders at the cellular and network levels. Mol Psychiatry 2012; 17: 1239–1253.
Brennand KJ, Gage FH . Modeling psychiatric disorders through reprogramming. Dis Models Mech 2012; 5: 26–32.
Brennand KJ, Gage FH . Concise review: the promise of human induced pluripotent stem cell-based studies of schizophrenia. Stem Cells 2011; 29: 1915–1922.
Fairchild PJ . The challenge of immunogenicity in the quest for induced pluripotency. Nat Rev Immunol 2010; 10: 868–875.
Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 2015; 47: 702–709.
Sullivan PF, Daly MJ, O'Donovan M . Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012; 13: 537–551.
Lichtenstein P, Bjork C, Hultman CM, Scolnick E, Sklar P, Sullivan PF . Recurrence risks for schizophrenia in a Swedish national cohort. Psychol Med 2006; 36: 1417–1425.
Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 2009; 373: 234–239.
Sullivan PF, Kendler KS, Neale MC . Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 2003; 60: 1187–1192.
Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427.
Cross-Disorder Group of the Psychiatric Genomics C Cross-Disorder Group of the Psychiatric Genomics C Lee SH Cross-Disorder Group of the Psychiatric Genomics C Ripke S Cross-Disorder Group of the Psychiatric Genomics C Neale BM Cross-Disorder Group of the Psychiatric Genomics C Faraone SV Cross-Disorder Group of the Psychiatric Genomics C Purcell SM et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45: 984–994.
Malhotra D, Sebat J . CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 2012; 148: 1223–1241.
Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry 2011; 168: 302–316.
Tansey KE, Rees E, Linden DE, Ripke S, Chambert KD, Moran JL et al. Common alleles contribute to schizophrenia in CNV carriers. Mol Psychiatry advance online publication, 22 September 2015; doi:10.1038/mp.2015.143 [e-pub ahead of print].
Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kähler A, Akterin S et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45: 1150–1159.
Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014; 506: 185–190.
Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014; 506: 179–184.
Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 2015; 87: 1215–1233.
Network, Pathway Analysis Subgroup of Psychiatric Network, Pathway Analysis Subgroup of Psychiatric Genomics C . Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 2015; 18: 199–209.
Loohuis LM, Vorstman JA, Ori AP, Staats KA, Wang T, Richards AL et al. Genome-wide burden of deleterious coding variants increased in schizophrenia. Nat Commun 2015; 6: 7501.
Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 2013; 154: 518–529.
Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 2012; 149: 525–537.
Takahashi K, Yamanaka S . Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–872.
Robinton DA, Daley GQ . The promise of induced pluripotent stem cells in research and therapy. Nature 2012; 481: 295–305.
Jones CA, Watson DJ, Fone KC . Animal models of schizophrenia. Br J Pharmacol 2011; 164: 1162–1194.
Provenzano G, Zunino G, Genovesi S, Sgado P, Bozzi Y . Mutant mouse models of autism spectrum disorders. Dis Markers 2012; 33: 225–239.
Harrison PJ, Law AJ . Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biol Psychiatry 2006; 60: 132–140.
Jaaro-Peled H . Gene models of schizophrenia: DISC1 mouse models. Prog Brain Res 2009; 179: 75–86.
Mei L, Xiong WC . Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci 2008; 9: 437–452.
Chen XW, Feng YQ, Hao CJ, Guo XL, He X, Zhou ZY et al. DTNBP1, a schizophrenia susceptibility gene, affects kinetics of transmitter release. J Cell Biol 2008; 181: 791–801.
Papaleo E, Russo L, Shaikh N, Cipolla L, Fantucci P, De Gioia L . Molecular dynamics investigation of cyclic natriuretic peptides: dynamic properties reflect peptide activity. J Mol Graph Model 2010; 28: 834–841.
O'Tuathaigh CM, Harte M, O'Leary C, O'Sullivan GJ, Blau C, Lai D et al. Schizophrenia-related endophenotypes in heterozygous neuregulin-1 'knockout' mice. Eur J Neurosci 2010; 31: 349–358.
Feng YQ, Zhou ZY, He X, Wang H, Guo XL, Hao CJ et al. Dysbindin deficiency in sandy mice causes reduction of snapin and displays behaviors related to schizophrenia. Schizophr Res 2008; 106: 218–228.
Liu WS, Pesold C, Rodriguez MA, Carboni G, Auta J, Lacor P et al. Down-regulation of dendritic spine and glutamic acid decarboxylase 67 expressions in the reelin haploinsufficient heterozygous reeler mouse. Proc Natl Acad Sci USA 2001; 98: 3477–3482.
Podhorna J, Didriksen M . The heterozygous reeler mouse: behavioural phenotype. Behav Brain Res 2004; 153: 43–54.
Krueger DD, Howell JL, Hebert BF, Olausson P, Taylor JR, Nairn AC . Assessment of cognitive function in the heterozygous reeler mouse. Psychopharmacology 2006; 189: 95–104.
Tueting P, Doueiri MS, Guidotti A, Davis JM, Costa E . Reelin down-regulation in mice and psychosis endophenotypes. Neurosci Biobehav Rev 2006; 30: 1065–1077.
Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O'Donovan MC et al. Evaluating historical candidate genes for schizophrenia. Mol Psychiatry 2015; 20: 555–562.
Yang C, Al-Aama J, Stojkovic M, Keavney B, Trafford A, Lako M et al. Concise review: cardiac disease modeling using induced pluripotent stem cells. Stem Cells 2015; 33: 2643–2651.
Liang P, Du J . Human induced pluripotent stem cell for modeling cardiovascular diseases. Reg Med Res 2014; 2: 4.
Curry EL, Moad M, Robson CN, Heer R . Using induced pluripotent stem cells as a tool for modelling carcinogenesis. World J Stem Cells 2015; 7: 461–469.
Nishi M, Akutsu H, Kudoh A, Kimura H, Yamamoto N, Umezawa A et al. Induced cancer stem-like cells as a model for biological screening and discovery of agents targeting phenotypic traits of cancer stem cell. Oncotarget 2014; 5: 8665–8680.
Zheng A, Li Y, Tsang SH . Personalized therapeutic strategies for patients with retinitis pigmentosa. Exp Opin Biol Ther 2015; 15: 391–402.
Wiley LA, Burnight ER, Songstad AE, Drack AV, Mullins RF, Stone EM et al. Patient-specific induced pluripotent stem cells (iPSCs) for the study and treatment of retinal degenerative diseases. Prog Retin Eye Res 2015; 44: 15–35.
Abdelalim EM, Bonnefond A, Bennaceur-Griscelli A, Froguel P . Pluripotent stem cells as a potential tool for disease modelling and cell therapy in diabetes. Stem Cell Rev 2014; 10: 327–337.
Lysy PA, Weir GC, Bonner-Weir S . Concise review: pancreas regeneration: recent advances and perspectives. Stem Cells Transl Med 2012; 1: 150–159.
Crook JM, Wallace G, Tomaskovic-Crook E . The potential of induced pluripotent stem cells in models of neurological disorders: implications on future therapy. Exp Rev Neurother 2015; 15: 295–304.
Peitz M, Jungverdorben J, Brustle O . Disease-specific iPS cell models in neuroscience. Curr Mol Med 2013; 13: 832–841.
Brennand KJ, Landek-Salgado MA, Sawa A . Modeling heterogeneous patients with a clinical diagnosis of schizophrenia with induced pluripotent stem cells. Biol Psychiatry 2014; 75: 936–944.
Merkle FT, Eggan K . Modeling human disease with pluripotent stem cells: from genome association to function. Cell Stem Cell 2013; 12: 656–668.
Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011; 473: 221–225.
Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med 2011; 17: 1657–1662.
Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 2014; 515: 414–418.
Prilutsky D, Palmer NP, Smedemark-Margulies N, Schlaeger TM, Margulies DM, Kohane IS . iPSC-derived neurons as a higher-throughput readout for autism: promises and pitfalls. Trends Mol Med 2014; 20: 91–104.
Iwamoto K, Kato T . Gene expression profiling in schizophrenia and related mental disorders. Neuroscientist 2006; 12: 349–361.
Topol A, Zhu S, Tran N, Simone A, Fang G, Brennand KJ et al. Signaling in human induced pluripotent stem cell neural progenitor cells derived from four schizophrenia patients. Biol Psychiatry 2015; 78: e29–e34.
Farra N, Zhang WB, Pasceri P, Eubanks JH, Salter MW, Ellis J . Rett syndrome induced pluripotent stem cell-derived neurons reveal novel neurophysiological alterations. Mol Psychiatry 2012; 17: 1261–1271.
Stevens HE, Mariani J, Coppola G, Vaccarino FM . Neurobiology meets genomic science: the promise of human-induced pluripotent stem cells. Dev Psychopathol 2012; 24: 1443–1451.
Vaccarino FM, Stevens HE, Kocabas A, Palejev D, Szekely A, Grigorenko EL et al. Induced pluripotent stem cells: a new tool to confront the challenge of neuropsychiatric disorders. Neuropharmacology 2011; 60: 1355–1363.
Vaccarino FM, Urban AE, Stevens HE, Szekely A, Abyzov A, Grigorenko EL et al. Annual research review: the promise of stem cell research for neuropsychiatric disorders. J Child Psychol Psychiatry 2011; 52: 504–516.
Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry 2015; 20: 361–368.
Li J, Ma ZH, Shi MY, Malty RH, Aoki H, Minic Z et al. Identification of human neuronal protein complexes reveals biochemical activities and convergent mechanisms of action in autism spectrum disorders. Cell Syst 2015; 1: 361–374.
Bidinosti M, Botta P, Kruttner S, Proenca CC, Stoehr N, Bernhard M et al. CLK2 inhibition ameliorates autistic features associated with SHANK3 deficiency. Science 2016; 351: 1199–1203.
Ebert DH, Greenberg ME . Activity-dependent neuronal signalling and autism spectrum disorder. Nature 2013; 493: 327–337.
Bernstein HG, Steiner J, Guest PC, Dobrowolny H, Bogerts B . Glial cells as key players in schizophrenia pathology: recent insights and concepts of therapy. Schizophr Res 2015; 161: 4–18.
Kerns D, Vong GS, Barley K, Dracheva S, Katsel P, Casaccia P et al. Gene expression abnormalities and oligodendrocyte deficits in the internal capsule in schizophrenia. Schizophr Res 2010; 120: 150–158.
Uranova NA, Vikhreva OV, Rachmanova VI, Orlovskaya DD . Ultrastructural alterations of myelinated fibers and oligodendrocytes in the prefrontal cortex in schizophrenia: a postmortem morphometric study. Schizophr Res Treat 2011; 2011: 325789.
Goudriaan A, de Leeuw C, Ripke S, Hultman CM, Sklar P, Sullivan PF et al. Specific glial functions contribute to schizophrenia susceptibility. Schizophr Bull 2014; 40: 925–935.
Martins-de-Souza D . Proteome and transcriptome analysis suggests oligodendrocyte dysfunction in schizophrenia. J Psychiatr Res 2010; 44: 149–156.
Duncan LE, Holmans PA, Lee PH, O'Dushlaine CT, Kirby AW, Smoller JW et al. Pathway analyses implicate glial cells in schizophrenia. PLoS One 2014; 9: e89441.
Bigelow LB, Nasrallah HA, Rauscher FP . Corpus callosum thickness in chronic schizophrenia. Br J Psychiatr 1983; 142: 284–287.
Williams MR, Hampton T, Pearce RK, Hirsch SR, Ansorge O, Thom M et al. Astrocyte decrease in the subgenual cingulate and callosal genu in schizophrenia. Eur Arch Psychiatr Clin Neurosci 2013; 263: 41–52.
Webster MJ, O'Grady J, Kleinman JE, Weickert CS . Glial fibrillary acidic protein mRNA levels in the cingulate cortex of individuals with depression, bipolar disorder and schizophrenia. Neuroscience 2005; 133: 453–461.
Steffek AE, McCullumsmith RE, Haroutunian V, Meador-Woodruff JH . Cortical expression of glial fibrillary acidic protein and glutamine synthetase is decreased in schizophrenia. Schizophr Res 2008; 103: 71–82.
Zhai J, Zhang Q, Cheng L, Chen M, Wang K, Liu Y et al. Risk variants in the S100B gene, associated with elevated S100B levels, are also associated with visuospatial disability of schizophrenia. Behav Brain Res 2011; 217: 363–368.
Shirts BH, Wood J, Yolken RH, Nimgaonkar VL . Association study of IL10, IL1beta, and IL1RN and schizophrenia using tag SNPs from a comprehensive database: suggestive association with rs16944 at IL1beta. Schizophr Res 2006; 88: 235–244.
Lencz T, Morgan TV, Athanasiou M, Dain B, Reed CR, Kane JM et al. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol Psychiatry 2007; 12: 572–580.
Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D et al. Common variants conferring risk of schizophrenia. Nature 2009; 460: 744–747.
Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N et al. Schizophrenia risk from complex variation of complement component 4. Nature 2016; 530: 177–183.
Benros ME, Nielsen PR, Nordentoft M, Eaton WW, Dalton SO, Mortensen PB . Autoimmune diseases and severe infections as risk factors for schizophrenia: a 30-year population-based register study. Am J Psychiatry 2011; 168: 1303–1310.
Harding MJ, McGraw HF, Nechiporuk A . The roles and regulation of multicellular rosette structures during morphogenesis. Development 2014; 141: 2549–2558.
Lim KC, Crino PB . Focal malformations of cortical development: new vistas for molecular pathogenesis. Neuroscience 2013; 252: 262–276.
Harrison PJ . Postmortem studies in schizophrenia. Dialogues Clin Neurosci 2000; 2: 349–357.
Benes FM . Building models for postmortem abnormalities in hippocampus of schizophrenics. Schizophr Res 2015; 167: 73–83.
Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 2015; 527: 95–99.
Nakai J, Ohkura M, Imoto K . A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol 2001; 19: 137–141.
Glover JC, Sato K, Momose-Sato Y . Using voltage-sensitive dye recording to image the functional development of neuronal circuits in vertebrate embryos. Dev Neurobiol 2008; 68: 804–816.
Homma R, Baker BJ, Jin L, Garaschuk O, Konnerth A, Cohen LB et al. Wide-field and two-photon imaging of brain activity with voltage- and calcium-sensitive dyes. Methods Mol Biol 2009; 489: 43–79.
Kraehenbuehl TP, Langer R, Ferreira LS . Three-dimensional biomaterials for the study of human pluripotent stem cells. Nat Methods 2011; 8: 731–736.
Shao Y, Sang J, Fu J . On human pluripotent stem cell control: The rise of 3D bioengineering and mechanobiology. Biomaterials 2015; 52: 26–43.
Zhang D, Pekkanen-Mattila M, Shahsavani M, Falk A, Teixeira AI, Herland A . A 3D Alzheimer's disease culture model and the induction of P21-activated kinase mediated sensing in iPSC derived neurons. Biomaterials 2014; 35: 1420–1428.
Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME et al. Cerebral organoids model human brain development and microcephaly. Nature 2013; 501: 373–379.
Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 2015; 12: 671–678.
Sasai Y . Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell 2013; 12: 520–530.
Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 2015; 162: 375–390.
Lu C, Chen Q, Zhou T, Bozic D, Fu Z, Pan JQ et al. Micro-electrode array recordings reveal reductions in both excitation and inhibition in cultured cortical neuron networks lacking Shank3. Mol Psychiatry 2016; 21: 159–168.
Heikkila TJ, Yla-Outinen L, Tanskanen JM, Lappalainen RS, Skottman H, Suuronen R et al. Human embryonic stem cell-derived neuronal cells form spontaneously active neuronal networks in vitro. Exp Neurol 2009; 218: 109–116.
Odawara A, Saitoh Y, Alhebshi AH, Gotoh M, Suzuki I . Long-term electrophysiological activity and pharmacological response of a human induced pluripotent stem cell-derived neuron and astrocyte co-culture. Biochem Biophys Res Commun 2014; 443: 1176–1181.
Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci USA 2015; 112: E2725–E2734.
MacLaren EJ, Charlesworth P, Coba MP, Grant SG . Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro. Mol Cell Neurosci 2011; 47: 93–99.
Uhlhaas PJ, Singer W . Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci 2010; 11: 100–113.
Liang G, Zhang Y . Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell 2013; 13: 149–159.
Martinez-Fernandez A, Nelson TJ, Terzic A . Nuclear reprogramming strategy modulates differentiation potential of induced pluripotent stem cells. J Cardiovas Transl Res 2011; 4: 131–137.
Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 2011; 13: 541–549.
Nityanandam A, Baldwin KK . Advances in reprogramming-based study of neurologic disorders. Stem Cells Dev 2015; 24: 1265–1283.
Wu H, Xu J, Pang ZP, Ge W, Kim KJ, Blanchi B et al. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc Natl Acad Sci USA 2007; 104: 13821–13826.
Ji J, Ng SH, Sharma V, Neculai D, Hussein S, Sam M et al. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells 2012; 30: 435–440.
Liu P, Kaplan A, Yuan B, Hanna JH, Lupski JR, Reiner O . Passage number is a major contributor to genomic structural variations in mouse iPSCs. Stem Cells 2014; 32: 2657–2667.
Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D . Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet 2014; 10: e1004432.
Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol 2015; 33: 1173–1181.
Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A et al. A comparison of non-integrating reprogramming methods. Nat Biotechnol 2015; 33: 58–63.
Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol 2015; 33: 1173–1181.
Thomas SM, Kagan C, Pavlovic BJ, Burnett J, Patterson K, Pritchard JK et al. Reprogramming LCLs to iPSCs results in recovery of donor-specific gene expression signature. PLoS Genet 2015; 11: e1005216.
Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA 2012; 109: 12770–12775.
Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell 2013; 12: 573–586.
Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 2013; 13: 691–705.
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153: 910–918.
Paull D, Sevilla A, Zhou H, Hahn AK, Kim H, Napolitano C et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat Methods 2015; 12: 885–892.
Hook V, Brennand KJ, Kim Y, Toneff T, Funkelstein L, Lee KC et al. Human iPSC neurons display activity-dependent neurotransmitter secretion: aberrant catecholamine levels in schizophrenia neurons. Stem Cell Rep 2014; 3: 531–538.
Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Rep 2014; 2: 295–310.
Hashimoto-Torii K, Torii M, Fujimoto M, Nakai A, El Fatimy R, Mezger V et al. Roles of heat shock factor 1 in neuronal response to fetal environmental risks and its relevance to brain disorders. Neuron 2014; 82: 560–572.
Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 2014; 15: 79–91.
Pedrosa E, Sandler V, Shah A, Carroll R, Chang C, Rockowitz S et al. Development of patient-specific neurons in schizophrenia using induced pluripotent stem cells. J Neurogenet 2011; 25: 88–103.
Zhao D, Lin M, Chen J, Pedrosa E, Hrabovsky A, Fourcade HM et al. MicroRNA profiling of neurons generated using induced pluripotent stem cells derived from patients with schizophrenia and schizoaffective disorder, and 22q11.2 Del. PLoS One 2015; 10: e0132387.
Paulsen Bda S, de Moraes Maciel R, Galina A, Souza da Silveira M, dos Santos Souza C, Drummond H et al. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant 2012; 21: 1547–1559.
Paulsen Bda S, Cardoso SC, Stelling MP, Cadilhe DV, Rehen SK . Valproate reverts zinc and potassium imbalance in schizophrenia-derived reprogrammed cells. Schizophr Res 2014; 154: 30–35.
Srikanth P, Han K, Callahan DG, Makovkina E, Muratore CR, Lalli MA et al. Genomic DISC1 disruption in hiPSCs alters Wnt signaling and neural cell fate. Cell Rep 2015; 12: 1414–1429.
Robicsek O, Karry R, Petit I, Salman-Kesner N, Muller FJ, Klein E et al. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry 2013; 18: 1067–1076.
Tian Y, Voineagu I, Pasca SP, Won H, Chandran V, Horvath S et al. Alteration in basal and depolarization induced transcriptional network in iPSC derived neurons from Timothy syndrome. Genome Med 2014; 6: 75.
Krey JF, Pasca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R et al. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci 2013; 16: 201–209.
Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A et al. Isolation of MECP2-null Rett syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet 2011; 20: 2103–2115.
Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 2010; 143: 527–539.
Kim KY, Hysolli E, Park IH . Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci USA 2011; 108: 14169–14174.
Ananiev G, Williams EC, Li H, Chang Q . Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS One 2011; 6: e25255.
Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Loven J et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell 2013; 13: 446–458.
Ricciardi S, Ungaro F, Hambrock M, Rademacher N, Stefanelli G, Brambilla D et al. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol 2012; 14: 911–923.
Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature 2013; 503: 267–271.
Urbach A, Bar-Nur O, Daley GQ, Benvenisty N . Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell 2010; 6: 407–411.
Bar-Nur O, Caspi I, Benvenisty N . Molecular analysis of FMR1 reactivation in fragile-X induced pluripotent stem cells and their neuronal derivatives. J Mol Cell Biol 2012; 4: 180–183.
Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One 2011; 6: e26203.
Liu J, Koscielska KA, Cao Z, Hulsizer S, Grace N, Mitchell G et al. Signaling defects in iPSC-derived fragile X premutation neurons. Hum Mol Genet 2012; 21: 3795–3805.
Germain ND, Chen PF, Plocik AM, Glatt-Deeley H, Brown J, Fink JJ et al. Gene expression analysis of human induced pluripotent stem cell-derived neurons carrying copy number variants of chromosome 15q11-q13.1. Mol Autism 2014; 5: 44.
Zeng L, Zhang P, Shi L, Yamamoto V, Lu W, Wang K . Functional impacts of NRXN1 knockdown on neurodevelopment in stem cell models. PLoS One 2013; 8: e59685.
Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S et al. Human neuropsychiatric disease modeling using conditional deletion reveals synaptic transmission defects caused by heterozygous mutations in NRXN1. Cell Stem Cell 2015; 17: 316–328.
Khattak S, Brimble E, Zhang W, Zaslavsky K, Strong E, Ross PJ et al. Human induced pluripotent stem cell derived neurons as a model for Williams-Beuren syndrome. Mol Brain 2015; 8: 77.
Qian K, Huang CT, Chen H, Blackbourn LWt, Chen Y, Cao J et al. A simple and efficient system for regulating gene expression in human pluripotent stem cells and derivatives. Stem Cells 2014; 32: 1230–1238.
Acknowledgements
MP and OB were supported by the EU (HEALTH-F4-2013-602278-NeuroStemCellRepair; FP7-HEALTH-2010-266753-SCR&Tox, COLIPA; IMI 115582-EBiSC; PHC-03-2015-COSYN), the German Federal Ministry of Education and Research (BMBF; 01ZX1314A-IntegraMent), the North Rhine Westphalian Ministry for Innovation, Science and Research (StemCellFactory #z1403ts007a), BONFOR and the Hertie Foundation. PFS was supported by Swedish Research Council (D0886501). Y-MS was supported by the Ministry of Health (15-31063A), Czech Republic. SD was supported by the KG Jebsen Foundation, the Research Council of Norway (#223273) and the South-East Norway Health Authority (#2014101). AJH was supported by the Wellcome Trust Strategic Award (WT100202/Z/12/Z). DP was supported by the Netherlands Organization for Scientific Research (NWO VICI 453-14-005). AF was supported by the Swedish foundation for strategic research (SSF IB13-0074), StratNeuro, StratRegen and Jeansson foundation. JCG was supported by the Research Council of Norway, the South-East Norway Regional Health Authority and the University of Oslo. All authors are members of European iPSC Consortium for Neuropsychiatric Disorders—EURICND
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
PowerPoint slides
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Falk, A., Heine, V., Harwood, A. et al. Modeling psychiatric disorders: from genomic findings to cellular phenotypes. Mol Psychiatry 21, 1167–1179 (2016). https://doi.org/10.1038/mp.2016.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2016.89
This article is cited by
-
Neurite outgrowth deficits caused by rare PLXNB1 mutation in pediatric bipolar disorder
Molecular Psychiatry (2023)
-
Copy number variants (CNVs): a powerful tool for iPSC-based modelling of ASD
Molecular Autism (2020)
-
Exploring the neuropsychiatric spectrum using high-content functional analysis of single-cell signaling networks
Molecular Psychiatry (2020)
-
The promises and challenges of human brain organoids as models of neuropsychiatric disease
Nature Medicine (2016)