Abstract
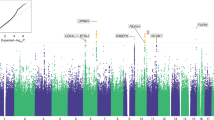
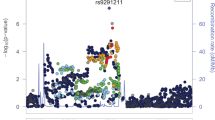
Opioid dependence, a severe addictive disorder and major societal problem, has been demonstrated to be moderately heritable. We conducted a genome-wide association study in Comorbidity and Trauma Study data comparing opioid-dependent daily injectors (N=1167) with opioid misusers who never progressed to daily injection (N=161). The strongest associations, observed for CNIH3 single-nucleotide polymorphisms (SNPs), were confirmed in two independent samples, the Yale-Penn genetic studies of opioid, cocaine and alcohol dependence and the Study of Addiction: Genetics and Environment, which both contain non-dependent opioid misusers and opioid-dependent individuals. Meta-analyses found five genome-wide significant CNIH3 SNPs. The A allele of rs10799590, the most highly associated SNP, was robustly protective (P=4.30E-9; odds ratio 0.64 (95% confidence interval 0.55–0.74)). Epigenetic annotation predicts that this SNP is functional in fetal brain. Neuroimaging data from the Duke Neurogenetics Study (N=312) provide evidence of this SNP’s in vivo functionality; rs10799590 A allele carriers displayed significantly greater right amygdala habituation to threat-related facial expressions, a phenotype associated with resilience to psychopathology. Computational genetic analyses of physical dependence on morphine across 23 mouse strains yielded significant correlations for haplotypes in CNIH3 and functionally related genes. These convergent findings support CNIH3 involvement in the pathophysiology of opioid dependence, complementing prior studies implicating the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) glutamate system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B et al. Familial transmission of substance use disorders. Arch Gen Psychiatry 1998; 55: 973–979.
Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry 1998; 55: 967–972.
Kendler KS, Jacobson KC, Prescott CA, Neale MC . Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry 2003; 160: 687–695.
Sun J, Bi J, Chan G, Oslin D, Farrer L, Gelernter J et al. Improved methods to identify stable, highly heritable subtypes of opioid use and related behaviors. Addict Behav 2012; 37: 1138–1144.
Agrawal A, Verweij KJ, Gillespie NA, Heath AC, Lessov-Schlaggar CN, Martin NG et al. The genetics of addiction-a translational perspective. Transl Psychiatry 2012; 2: e140.
Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL et al. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addict Biol 2014; 19: 111–121.
Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand F et al. ANKK1 TTC12, and NCAM1 polymorphisms and heroin dependence: importance of considering drug exposure. JAMA Psychiatry 2013; 70: 325–333.
Peng S, Du J, Jiang H, Fu Y, Chen H, Sun H et al. The dopamine receptor D1 gene is associated with the length of interval between first heroin use and onset of dependence in Chinese Han heroin addicts. J Neural Transm 2013; 120: 1591–1598.
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661–678.
Nutt D, King LA, Saulsbury W, Blakemore C . Development of a rational scale to assess the harm of drugs of potential misuse. Lancet 2007; 369: 1047–1053.
Linares OA . The Linares addictive potential model. J Pharm Nutr Sci 2012; 2: 132–139.
Anthony JC, Warner LA, Kessler RC . Comparative epidemiology of dependence on tobacco, alcohol, controlled substances, and inhalants basic findings From the National Comorbidity Survey. Exp Clin Psychopharmacol 1994; 2: 244–268.
Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PAF et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 2007; 16: 36–49.
Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008; 452: 638–642.
Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet 2010; 42: 436–440.
Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 2010; 42: 448–453.
Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 2010; 42: 441–447.
Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry 2012; 17: 445–450.
Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 2014; 19: 41–49.
Shand FL, Degenhardt L, Slade T, Nelson EC . Sex differences amongst dependent heroin users: histories, clinical characteristics and predictors of other substance dependence. Addict Behav 2011; 36: 27–36.
Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA 2010; 107: 5082–5087.
Bennett SN, Caporaso N, Fitzpatrick AL, Agrawal A, Barnes K, Boyd HA et al. Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience. Genet Epidemiol 2011; 35: 159–173.
Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H et al. Genome-wide association study of opioid cependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry 2014; 76: 66–74.
Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR et al. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry 2014; 19: 717–723.
Okvist A, Fagergren P, Whittard J, Garcia-Osta A, Drakenberg K, Horvath MC et al. Dysregulated postsynaptic density and endocytic zone in the amygdala of human heroin and cocaine abusers. Biol Psychiatry 2011; 69: 245–252.
Cai YQ, Wang W, Hou YY, Zhang Z, Xie J, Pan ZZ . Central amygdala GluA1 facilitates associative learning of opioid reward. J Neurosci 2013; 33: 1577–1588.
Van den Oever MC, Goriounova NA, Li KW, Van der Schors RC, Binnekade R, Schoffelmeer AN et al. Prefrontal cortex AMPA receptor plasticity is crucial for cue-induced relapse to heroin-seeking. Nat Neurosci 2008; 11: 1053–1058.
Xia Y, Portugal GS, Fakira AK, Melyan Z, Neve R, Lee HT et al. Hippocampal GluA1-containing AMPA receptors mediate context-dependent sensitization to morphine. J Neurosci 2011; 31: 16279–16291.
Aitta-aho T, Möykkynen TP, Panhelainen AE, Vekovischeva OY, Bäckström P, Korpi ER . Importance of GluA1 subunit-containing AMPA glutamate receptors for morphine state-dependency. PLoS One 2012; 7: e38325.
Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI Jr. et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol 1994; 55: 149–158.
Patterson N, Price AL, Reich D . Population structure and eigenanalysis. PLoS Genet 2006; 2: e190.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Willer CJ, Li Y, Abecasis GR . METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191.
So HC, Gui AH, Cherny SS, Sham PC . Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol 2011; 35: 310–317.
Hall WD, Ross JE, Lynskey MT, Law MG, Degenhardt LJ . How many dependent heroin users are there in Australia? Med J Aust 2000; 173: 528–531.
Degenhardt L, Hall W . The extent of illicit drug use and dependence, and their contribution to the global burden of disease. Lancet 2012; 379: 55–70.
Whalen PJ, Phelps EA (eds). The Human Amygdala. Guilford Press: New York, NY, USA, 2009.
Benjamini Y, Hochberg Y . Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 1995; 57: 289–300.
Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D et al. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J Neurosci 2012; 32: 17454–17464.
Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA et al. The Human Epigenome Browser at Washington University. Nat Methods 2011; 8: 989–990.
Zhou X, Lowdon RF, Li D, Lawson HA, Madden PAF, Costello JF et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods 2013; 10: 375–376.
Zhou X, Li D, Zhang B, Lowdon RF, Rockweiler NB, Sears RL et al. Epigenomic annotation of genetic variants using the Roadmap Epigenome Browser. Nat Biotechnol 2015; 33: 345–346.
Yeh SH, Mao SC, Lin HC, Gean PW . Synaptic expression of glutamate receptor after encoding of fear memory in the rat amygdala. Mol Pharmacol 2006; 69: 299–308.
Dalton GL, Wang YT, Floresco SB, Phillips AG . Disruption of AMPA receptor endocytosis impairs the extinction, but not acquisition of learned fear. Neuropsychopharmacology 2008; 33: 2416–2426.
Nedelescu H, Kelso CM, Lázaro-Muñoz G, Purpura M, Cain CK, Ledoux JE et al. Endogenous GluR1-containing AMPA receptors translocate to asymmetric synapses in the lateral amygdala during the early phase of fear memory formation: an electron microscopic immunocytochemical study. J Comp Neurol 2010; 518: 4723–4739.
Nikolova YS, Koenen KC, Galea S, Wang CM, Seney ML, Sibille E et al. Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci 2014; 17: 1153–1155.
Plichta MM, Grimm O, Morgen K, Mier D, Sauer C, Haddad L et al. Amygdala habituation: a reliable fMRI phenotype. Neuroimage 2014; 103C: 383–390.
Chu LF, Liang DY, Li X, Sahbaie P, D'arcy N, Liao G et al. From mouse to man: the 5-HT3 receptor modulates physical dependence on opioid narcotics. Pharmacogenet Genomics 2009; 19: 193–205.
Zheng M, Dill D, Peltz G . A better prognosis for genetic association studies in mice. Trends Genet 2012; 28: 62–69.
Liang DY, Zheng M, Sun Y, Sahbaie P, Low SA, Peltz G et al. The Netrin-1 receptor DCC is a regulator of maladaptive responses to chronic morphine administration. BMC Genomics 2014; 15: 345.
Schwenk J, Baehrens D, Haupt A, Bildl W, Boudkkazi S, Roeper J et al. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron 2014; 84: 41–54.
Sumioka A, Brown TE, Kato AS, Bredt DS, Kauer JA, Tomita S. PDZ . binding of TARPγ-8 controls synaptic transmission but not synaptic plasticity. Nat Neurosci 2011; 14: 1410–1412.
Harmel N, Cokic B, Zolles G, Berkefeld H, Mauric V, Fakler B et al. AMPA receptors commandeer an ancient cargo exporter for use as an auxiliary subunit for signaling. PLoS One 2012; 7: e30681.
Herring BE, Shi Y, Suh YH, Zheng CY, Blankenship SM, Roche KW et al. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron 2013; 77: 1083–1096.
Coombs ID, Soto D, Zonouzi M, Renzi M, Shelley C, Farrant M et al. Cornichons modify channel properties of recombinant and glial AMPA receptors. J Neurosci 2012; 32: 9796–9804.
Boudkkazi S, Brechet A, Schwenk J, Fakler B . Cornichon2 dictates the time course of excitatory transmission at individual hippocampal synapses. Neuron 2014; 82: 848–858.
Shanks NF, Cais O, Maruo T, Savas JN, Zaika EI, Azumaya CM et al. Molecular dissection of the interaction between the AMPA receptor and cornichon homolog-3. J Neurosci 2014; 34: 12104–12120.
Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E et al. GTEx Consortium, The genotype-tissue expression (GTEx) project. Nat Genet 2013; 45: 580–585.
Duffy DL, Iles MM, Glass D, Zhu G, Barrett JH, Höiom V et al. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am J Hum Genet 2010; 87: 6–16.
Gunduz-Cinar O, MacPherson KP, Cinar R, Gamble-George J, Sugden K, Williams B et al. Convergent translational evidence of a role for anandamide in amygdala-mediated fear extinction, threat processing and stress-reactivity. Mol Psychiatry 2013; 18: 813–823.
Day CA, Ross J, Dietze P, Dolan K . Initiation to heroin injecting among heroin users in Sydney, Australia: cross sectional survey. Harm Reduct J 2005; 2: 2.
Des Jarlais DC, Arasteh K, Perlis T, Hagan H, Heckathorn DD, Mcknight C et al. The transition from injection to non-injection drug use: long-term outcomes among heroin and cocaine users in New York City. Addiction 2007; 102: 778–785.
Acknowledgements
Funding support for the CATS was provided by the National Institute on Drug Abuse (R01 DA17305); GWAS genotyping services at the CIDR at The Johns Hopkins University were supported by the National Institutes of Health (contract N01-HG-65403). Funding for the Yale-Penn genetic studies of opioid, cocaine, or alcohol dependence was provided by the National Institutes of Health (RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, 5T32GM007205-38, 8UL1TR000142-07); and the Veterans Affairs Connecticut and Veterans Affairs Philadelphia Mental Illness Research, Education and Clinical Centers. Genotyping services at the CIDR at The Johns Hopkins University were supported by the National Institutes of Health (contract N01-HG-65403). Funding support for the SAGE was provided through the NIH Genes, Environment and Health Initiative (GEI) (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of data sets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423, R01 DA019963). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (HHSN268200782096C). The Duke Neurogenetics Study is supported by Duke University and National Institute on Drug Abuse grant DA033369. AA also receives support from R01 DA23668 and K02 DA32573. GWM and NRW are supported by NHMRC Fellowships. TW, BZ and XZ receive support from DA027995, R01HG007354, R01HG007175, R01ES024992 and ACS grant RSG-14-049-01-DMC. ARH receives support through National Institute on Drug Abuse grants DA033369 and DA031579. RB receives support from the Klingenstein Third Generation Foundation and the National Institutes of Health (NIA R01-AG045231). CHD receives support from T32DA007313. CEC receives support from NSF DGE-1143954.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Although unrelated to the current study, Dr Kranzler has been a consultant or advisory board member for Alkermes, Lilly, Lundbeck, Pfizer and Roche. He is also a member of the American Society of Clinical Psychopharmacology's Alcohol Clinical Trials Initiative, which is supported by Lilly, Lundbeck, Abbott and Pfizer. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information is available at the Molecular Psychiatry website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Nelson, E., Agrawal, A., Heath, A. et al. Evidence of CNIH3 involvement in opioid dependence. Mol Psychiatry 21, 608–614 (2016). https://doi.org/10.1038/mp.2015.102
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2015.102
This article is cited by
-
SNP-based and haplotype-based genome-wide association on drug dependence in Han Chinese
BMC Genomics (2024)
-
Methamphetamine-induced region-specific transcriptomic and epigenetic changes in the brain of male rats
Communications Biology (2023)
-
A genome-wide association, polygenic risk score and sex study on opioid use disorder treatment outcomes
Scientific Reports (2023)
-
Genetics and prescription opioid use (GaPO): study design for consenting a cohort from an existing biobank to identify clinical and genetic factors influencing prescription opioid use and abuse
BMC Medical Genomics (2021)
-
The Genomics of Opioid Addiction Longitudinal Study (GOALS): study design for a prospective evaluation of genetic and non-genetic factors for development of and recovery from opioid use disorder
BMC Medical Genomics (2021)