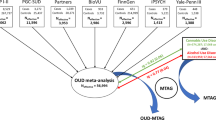
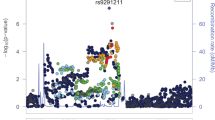
Abstract
Despite an estimated heritability of ~50%, genome-wide association studies of opioid use disorder (OUD) have revealed few genome-wide significant loci. We conducted a cross-ancestry meta-analysis of OUD in the Million Veteran Program (N = 425,944). In addition to known exonic variants in OPRM1 and FURIN, we identified intronic variants in RABEPK, FBXW4, NCAM1 and KCNN1. A meta-analysis including other datasets identified a locus in TSNARE1. In total, we identified 14 loci for OUD, 12 of which are novel. Significant genetic correlations were identified for 127 traits, including psychiatric disorders and other substance use-related traits. The only significantly enriched cell-type group was CNS, with gene expression enrichment in brain regions previously associated with substance use disorders. These findings increase our understanding of the biological basis of OUD and provide further evidence that it is a brain disease, which may help to reduce stigma and inform efforts to address the opioid epidemic.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The full summary-level association data from the meta-analysis are available through dbGaP at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.
Code availability
Imputation was performed using Minimac3 (https://genome.sph.umich.edu/wiki/Minimac3). GWAS was performed using PLINK2 (https://www.cog-genomics.org/plink2). Meta-analyses were performed using METAL (https://genome.sph.umich.edu/wiki/METAL_Documentation). GCTA (https://cnsgenomics.com/software/gcta/#Overview) was used for identification of independent loci (GCTA-COJO) and gene-based analysis (GCTA-fastBAT). FUMA (https://fuma.ctglab.nl/) was used for gene association, functional enrichment and gene-set enrichment analyses. Transcriptomic analyses were performed using S-PrediXcan and S-MultiXcan (https://github.com/hakyimlab/MetaXcan). LDSC (https://github.com/bulik/ldsc) was used for heritability estimation, genetic correlation analysis (also using the CTG-VL; https://genoma.io) and heritability enrichment analyses. Trans-ancestry genetic correlation was estimated using Popcorn (https://github.com/brielin/Popcorn). PRS analyses were performed using PRS-CS (https://github.com/getian107/PRScs). PheWAS analyses were run using the PheWAS R package (https://github.com/PheWAS/PheWAS). The MendelianRandomization R package (https://cran.r-project.org/web/packages/MendelianRandomization/index.html) was used for MR analyses. Genomic SEM was conducted using the GenomicsSEM R package (https://github.com/GenomicSEM/GenomicSEM).
References
American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders (American Psychiatric Association, 2013); https://doi.org/10.1176/appi.books.9780890425596
Strang, J. et al. Opioid use disorder. Nat. Rev. Dis. Primers 6, 3 (2020).
Vowles, K. E. et al. Rates of opioid misuse, abuse, and addiction in chronic pain: a systematic review and data synthesis. Pain 156, 569–576 (2015).
Centers for Disease Control and Prevention Vital signs: overdoses of prescription opioid pain relievers and other drugs among women–United States, 1999–2010. MMWR Morb. Mortal. Wkly Rep. 62, 537–542 (2013).
Key Substance Use and Mental Health Indicators in the United States: Results from the 2019 National Survey on Drug Use and Health (Substance Abuse and Mental Health Services Administration, 2020); https://www.samhsa.gov/data/sites/default/files/reports/rpt29393/2019NSDUHFFRPDFWHTML/2019NSDUHFFR090120.htm
Wilson, N. Drug and opioid-involved overdose deaths—United States, 2017–2018. MMWR Morb. Mortal. Wkly Rep. 69, 290–297 (2020).
Kendler, K. S., Jacobson, K. C., Prescott, C. A. & Neale, M. C. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am. J. Psychiatry 160, 687–695 (2003).
Gelernter, J. et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol. Psychiatry 76, 66–74 (2014).
Cheng, Z. et al. Genome-wide association study identifies a regulatory variant of rgma associated with opioid dependence in European Americans. Biol. Psychiatry 84, 762–770 (2018).
Nelson, E. C. et al. Evidence of CNIH3 involvement in opioid dependence. Mol. Psychiatry 21, 608–614 (2016).
Polimanti, R. et al. Leveraging genome-wide data to investigate differences between opioid use vs. opioid dependence in 41,176 individuals from the Psychiatric Genomics Consortium. Mol. Psychiatry 25, 1673–1687 (2020).
Song, W. et al. Genome-wide association analysis of opioid use disorder: a novel approach using clinical data. Drug Alcohol Depend. 217, 108276 (2020).
Zhou, H. et al. Association of OPRM1 functional coding variant with opioid use disorder: a genome-wide association study. JAMA Psychiatry 77, 1072–1080 (2020).
Deak, J. D. et al. Genome-wide association study and multi-trait analysis of opioid use disorder identifies novel associations in 639,709 individuals of European and African ancestry. Mol. Psychiatry https://doi.org/10.1038/s41380-022-01709-1 (2022).
Sanchez-Roige, S. et al. Genome-wide association study of problematic opioid prescription use in 132,113 23andMe research participants of European ancestry. Mol. Psychiatry https://doi.org/10.1038/s41380-021-01335-3 (2021).
Gaddis, N. et al. Multi-trait genome-wide association study of opioid addiction: OPRM1 and beyond. Preprint at medRxiv https://doi.org/10.1101/2021.09.13.21263503v1 (2021).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
The GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Davydov, E. V. et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 6, e1001025 (2010).
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Barbeira, A. N. et al. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 15, e1007889 (2019).
Qi, T. et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 9, 2282 (2018).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Cuellar-Partida, G. et al. Complex-Trait Genetics Virtual Lab: a community-driven web platform for post-GWAS analyses. Preprint at bioRxiv https://doi.org/10.1101/518027 (2019).
Crist, R. C. & Berrettini, W. H. Pharmacogenetics of OPRM1. Pharmacol. Biochem. Behav. 123, 25–33 (2014).
Crist, R. C., Reiner, B. C. & Berrettini, W. H. A review of opioid addiction genetics. Curr. Opin. Psychol. 27, 31–35 (2019).
Moningka, H., Lichenstein, S. & Yip, S. W. Current understanding of the neurobiology of opioid use disorder: an overview. Curr. Behav. Neurosci. Rep. 6, 1–11 (2019).
Gelernter, J., Kranzler, H. & Cubells, J. Genetics of two mu opioid receptor gene (OPRM1) exon I polymorphisms: population studies, and allele frequencies in alcohol- and drug-dependent subjects. Mol. Psychiatry 4, 476–483 (1999).
Howe, K. L. et al. Ensembl 2021. Nucleic Acids Res. 49, D884–D891 (2021).
Zhang, H. et al. Association between two µ-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum. Mol. Genet. 15, 807–819 (2006).
Breslin, M. B. et al. Differential processing of proenkephalin by prohormone convertases 1(3) and 2 and furin. J. Biol. Chem. 268, 27084–27093 (1993).
Christakoudi, S., Evangelou, E., Riboli, E. & Tsilidis, K. K. GWAS of allometric body-shape indices in UK Biobank identifies loci suggesting associations with morphogenesis, organogenesis, adrenal cell renewal and cancer. Sci. Rep. 11, 10688 (2021).
Liu, M. et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019).
Grice, D. E. et al. Transcriptional profiling of C57 and DBA strains of mice in the absence and presence of morphine. BMC Genomics 8, 76 (2007).
Yu, L. et al. Activity in projection neurons from prelimbic cortex to the PVT is necessary for retrieval of morphine withdrawal memory. Cell Rep. 35, 108958 (2021).
Fujita-Hamabe, W., Nakamoto, K. & Tokuyama, S. Involvement of NCAM and FGF receptor signaling in the development of analgesic tolerance to morphine. Eur. J. Pharmacol. 672, 77–82 (2011).
Yang, B.-Z. et al. Association of haplotypic variants in DRD2, ANKK1, TTC12 and NCAM1 to alcohol dependence in independent case control and family samples. Hum. Mol. Genet. 16, 2844–2853 (2007).
Yang, B.-Z. et al. Haplotypic variants in DRD2, ANKK1, TTC12 and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol. Clin. Exp. Res. 32, 2117–2127 (2008).
Pasman, J. A. et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat. Neurosci. 21, 1161–1170 (2018).
Vujkovic, M. et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 52, 680–691 (2020).
Lee, J. J. et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018).
Amare, A. T. et al. Bivariate genome-wide association analyses of the broad depression phenotype combined with major depressive disorder, bipolar disorder or schizophrenia reveal eight novel genetic loci for depression. Mol. Psychiatry 25, 1420–1429 (2020).
Koob, G. F. Neurobiology of opioid addiction: opponent process, hyperkatifeia and negative reinforcement. Biol. Psychiatry 87, 44–53 (2020).
Leshner, A. I. Addiction is a brain disease, and it matters. Science 278, 45–47 (1997).
Karlsson Linnér, R. et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat. Neurosci. 24, 1367–1376 (2021).
Vanyukov, M. M. et al. Common liability to addiction and ‘gateway hypothesis’: Theoretical, empirical and evolutionary perspective. Drug Alcohol Depend. 123, S3–S17 (2012).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic relationships, novel loci and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482 (2019).
Howell, B. A. et al. Validity of incident opioid use disorder (OUD) diagnoses in administrative data: a chart verification study. J. Gen. Intern. Med. 36, 1264–1270 (2021).
Gaziano, J. M. et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 70, 214–223 (2016).
Delaneau, O., Zagury, J.-F., Robinson, M. R., Marchini, J. L. & Dermitzakis, E. T. Accurate, scalable and integrative haplotype estimation. Nat. Commun. 10, 5436 (2019).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Fang, H. et al. Harmonizing genetic ancestry and self-identified race/ethnicity in genome-wide association studies. Am. J. Hum. Genet. 105, 763–772 (2019).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Altshuler, D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018).
Bernstein, B. E. et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 (2010).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Bakshi, A. et al. Fast set-based association analysis using summary data from GWAS identifies novel gene loci for human complex traits. Sci. Rep. 6, 32894 (2016).
Barbeira, A. N. et al. Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biol. 22, 49 (2021).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Aguet, F. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Fromer, M. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 19, 1442–1453 (2016).
Ng, B. et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 20, 1418–1426 (2017).
Freshour, S. L. et al. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 49, D1144–D1151 (2021).
Brown, B. C., Asian Genetic Epidemiology Network Type 2 Diabetes Consortium, Ye, C. J., Price, A. L. & Zaitlen, N. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet. 99, 76–88 (2016).
Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Dennis, J. K. et al. Clinical laboratory test-wide association scan of polygenic scores identifies biomarkers of complex disease. Genome Med. 13, 6 (2021).
Grotzinger, A. D. et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 3, 513–525 (2019).
Zhou, H. et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat. Neurosci. 23, 809–818 (2020).
Johnson, E. C. et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry 7, 1032–1045 (2020).
Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Yu, D. et al. Interrogating the genetic determinants of Tourette’s syndrome and other tic disorders through genome-wide association studies. Am. J. Psychiatry 176, 217–227 (2019).
International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS). Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol. Psychiatry 23, 1181–1188 (2018).
Acknowledgements
This work was supported by Merit Review Awards from the US Department of Veterans Affairs Biomedical Laboratory Research and Development Service (no. I01 BX003341 (to A.C.J. and H.R.K.)) and Clinical Science Research and Development Service (no. I01 CX001734 (to K.M.K.)); the VISN 4 Mental Illness Research, Education and Clinical Center (to H.R.K.); NIAAA grant K01 AA028292 (to R.L.K.); and NIDA grant DA046345 (to H.R.K.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed in this article are those of the authors and do not necessarily represent the position or policy of the Department of Veterans Affairs or the US Government.
Author information
Authors and Affiliations
Consortia
Contributions
R.L.K., H.X. and H.R.K. conceived analyses. R.L.K. and H.R.K. wrote the first draft and prepared all drafts for submission; R.L.K. accomplished primary analyses; H.X., S.T., M.N. and H.Z. conducted additional analyses; R.L.K., L.K.D., S.S.-R. and J.G. supervised additional analyses; R.V.-S., E.E.H., C.T.R., M.N., L.K.D. and S.S.-R. provided critical support regarding phenotypes and data in individual datasets; and K.M.K., A.C.J. and H.R.K. provided resource support. All authors reviewed the manuscript and approved it for submission.
Corresponding author
Ethics declarations
Competing interests
H.R.K. is a member of advisory boards for Dicerna Pharmaceuticals, Sophrosyne Pharmaceuticals and Enthion Pharmaceuticals; a consultant to Sobrera Pharmaceuticals; the recipient of research funding and medication supplies for an investigator-initiated study from Alkermes, and a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by Alkermes, Dicerna, Ethypharm, Lundbeck, Mitsubishi and Otsuka. J.G. and H.R.K. are holders of US patent no. 10,900,082 titled: ‘Genotype-guided dosing of opioid agonists,’ issued 26 January 2021. The other authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Ditte Demontis and Andrew McIntosh for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Material and Figs. 1–9
Rights and permissions
About this article
Cite this article
Kember, R.L., Vickers-Smith, R., Xu, H. et al. Cross-ancestry meta-analysis of opioid use disorder uncovers novel loci with predominant effects in brain regions associated with addiction. Nat Neurosci 25, 1279–1287 (2022). https://doi.org/10.1038/s41593-022-01160-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01160-z
This article is cited by
-
Medical and genetic correlates of long-term buprenorphine treatment in the electronic health records
Translational Psychiatry (2024)
-
Pleiotropy and genetically inferred causality linking multisite chronic pain to substance use disorders
Molecular Psychiatry (2024)
-
Multi-ancestry meta-analysis of tobacco use disorder identifies 461 potential risk genes and reveals associations with multiple health outcomes
Nature Human Behaviour (2024)
-
Multi-trait genome-wide association analyses leveraging alcohol use disorder findings identify novel loci for smoking behaviors in the Million Veteran Program
Translational Psychiatry (2023)
-
The Genetically Informed Neurobiology of Addiction (GINA) model
Nature Reviews Neuroscience (2023)