Abstract
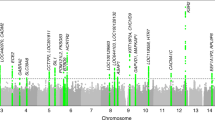
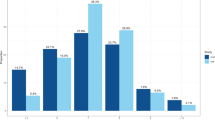
Usual sleep duration is a heritable trait correlated with psychiatric morbidity, cardiometabolic disease and mortality, although little is known about the genetic variants influencing this trait. A genome-wide association study (GWAS) of usual sleep duration was conducted using 18 population-based cohorts totaling 47 180 individuals of European ancestry. Genome-wide significant association was identified at two loci. The strongest is located on chromosome 2, in an intergenic region 35- to 80-kb upstream from the thyroid-specific transcription factor PAX8 (lowest P=1.1 × 10−9). This finding was replicated in an African-American sample of 4771 individuals (lowest P=9.3 × 10−4). The strongest combined association was at rs1823125 (P=1.5 × 10−10, minor allele frequency 0.26 in the discovery sample, 0.12 in the replication sample), with each copy of the minor allele associated with a sleep duration 3.1 min longer per night. The alleles associated with longer sleep duration were associated in previous GWAS with a more favorable metabolic profile and a lower risk of attention deficit hyperactivity disorder. Understanding the mechanisms underlying these associations may help elucidate biological mechanisms influencing sleep duration and its association with psychiatric, metabolic and cardiovascular disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Partinen M, Kaprio J, Koskenvuo M, Putkonen P, Langinvainio H . Genetic and environmental determination of human sleep. Sleep 1983; 6: 179–185.
Heath AC, Kendler KS, Eaves LJ, Martin NG . Evidence for genetic influences on sleep disturbance and sleep pattern in twins. Sleep 1990; 13: 318–335.
Watson NF, Buchwald D, Vitiello MV, Noonan C, Goldberg J . A twin study of sleep duration and body mass index. J Clin Sleep Med 2010; 6: 11–17.
Sehgal A, Mignot E . Genetics of sleep and sleep disorders. Cell 2011; 146: 194–207.
Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2001; 291: 1040–1043.
Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 2005; 434: 640–644.
He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL Jr et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science 2009; 325: 866–870.
Allebrandt KV, Teder-Laving M, Akyol M, Pichler I, Müller-Myhsok B, Pramstaller P et al. CLOCK gene variants associate with sleep duration in two independent populations. Biol Psychiatry 2010; 67: 1040–1047.
Evans DS, Parimi N, Nievergelt CM, Blackwell T, Redline S, Ancoli-Israel S et al. Common genetic variants in ARNTL and NPAS2 and at chromosome 12p13 are associated with objectively measured sleep traits in the elderly. Sleep 2013; 36: 431–446.
Utge S, Kronholm E, Partonen T, Soronen P, Ollila HM, Loukola A et al2011 Shared genetic background for regulation of mood and sleep: association of GRIA3 with sleep duration in healthy Finnish women. Sleep 34: 1309–1316.
Gottlieb DJ, O'Connor GT, Wilk JB . Genome-wide association of sleep and circadian phenotypes. BMC Med Genet 2007; 8 (Suppl 1): S9.
Allebrandt KV, Amin N, Müller-Myhsok B, Esko T, Teder-Laving M, Azevedo RV et al. A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol Psychiatry 2011; 18: 122–132.
Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009; 2: 73–80.
The ARIC investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol 1989; 129: 687–702.
Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol 1991; 1: 263–276.
Dawber TR, Meadors GF, Moore FE . Epidemiological approaches to heart disease: the Framingham study. Am J Public Health 1951; 41: 279–286.
Feinleib M, Kannel W, Garrison R, McNamara P, Castelli W . The Framingham Offspring Study: design and preliminary data. Prev Med 1975; 4: 518–525.
Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ et al. The Third Generation Cohort of the National Heart Lung and Blood Institute’s Framingham Heart Study: design recruitment and initial examination. Am J Epidemiol 2007; 165: 1328–1335.
Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL et al. New loci associated with kidney function and chronic kidney disease. Nat Genet 2010; 42: 376–384.
Barker DJP, Osmond C, Forsen TJ, Kajantie E, Eriksson JG . Trajectories of growth among children who have coronary events as adults. N Engl J Med 2005; 353: 1802–1809.
Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, Rafferty I et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet 2008; 4, p e1000072.
Blank JB, Cawthon PM, Carrion-Petersen ML, Harper L, Johnson JP, Mitson E et al. Overview of recruitment for the osteoporotic fractures in men study (MrOS). Contemp Clin Trials 2005; 26: 557–568.
Orwoll E, Blank JB, Barrett-Connor E, Cauley J, Cummings S, Ensrud K et al. Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study—a large observational study of the determinants of fracture in older men. Contemp Clin Trials 2005; 26: 569–585.
Bouchard C . Genetic epidemiology association and sib-pair linkage: results from the Quebec Family Study In Bray GA, Ryan DH (eds) Molecular and Genetic Aspects of Obesity. Louisiana State University Press: Baton Rouge, LA, USA, pp 470–481, 1996.
Hofman A, van Duijn CM, Franco OH, Ikram MA, Janssen HL, Klaver CC et al. The Rotterdam Study: 2012 objectives and design update. Eur J Epidemiol 2011; 26: 657–686.
Völzke H, Alte D, Schmidt CO, Radke D, Lorbeer R et al. Study of Health in Pomerania (SHIP) – a community cohort and repeated survey approach to comprehensively assess main health determinants among the general adult population. Int J Epidemiol 2011; 40: 294–307.
Cummings SR, Nevitt MC, Browner WS, Stone K, Fox KM, Ensrud KE et al. Risk factors for hip fracture in white women. Study of Osteoporotic Fractures Research Group. N Engl J Med 1995; 332: 767–773.
Moayyeri A, Hammond CJ, Valdes AM, Spector TD . Cohort Profile: TwinsUK and Healthy Ageing Twin Study. Int J Epidemiol 2012; 42: 76–85.
Young T, Palta M, Dempsey J, Skatrud J, Weber S, Badr S . The occurrence of sleep-disordered breathing among middle-aged adults. N Engl J Med 1993; 328: 1230–1235.
Raitakari OT, Juonala M, Rönnemaa T, Keltikangas-Järvinen L, Räsänen L, Pietikäinen M et al. Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol 2008; 37: 1220–1226.
Rimm EB, Giovannucci EL, Willett WC, Colditz GA, Ascherio A, Rosner B et al. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet 1991; 338: 464–468.
Colditz GA, Hankinson SE . The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer 2005; 5: 388–396.
Musunuru K, Lettre G, Young T, Farlow DN, Pirruccello JP, Ejebe KG et al. Candidate gene association resource (CARe): design methods and proof of concept. Circ Cardiovasc Genet 2010; 3: 267–275.
Almasy L, Blangero J . Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998; 62: 1198–1211.
Willer C, Li Y, Abecasis G . METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191.
Yang J, Ferreira T, Morris AP, Medland SE Genetic Investigation of ANthropometric Traits (GIANT) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Madden PA, Heath AC et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012; 44: 369–375, S1-3.
Gauderman WJ, Morrison JM QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. http://hydra.usc.edu/gxe, 2006.
Johnson AD, Handsaker RE, Pulit S, Nizzari MM, O’Donnell CJ, de Bakker PIW . SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008; 24: 2938–2939.
Porcu E, Medici M, Pistis G, Volpato CB, Wilson SG, Cappola AR et al. A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet 2013; 9: e1003266.
Nitsch R, Dato VD, Gennaro AD, Cristofaro TD, Abbondante S, De Felice M et al. Comparative genomics reveals a functional thyroid-specific element in the far upstream region of the PAX8 gene. Genomics 2010; 11: 306.
Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 2012; 44: 1084–1089.
Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W et al. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet 2010; 87: 779–789.
Morselli L, Leproult R, Balbo M, Spiegel K . Role of sleep duration in the regulation of glucose metabolism and appetite. Best Pract Res Clin Endocrinol Metab 2010; 24: 687–702.
Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010; 42: 105–116.
Soranzo N, Sanna S, Wheeler E, Gieger C, Radke D, Dupuis J et al. Common variants at 10 genomic loci influence hemoglobin A1C levels via glycemic and nonglycemic pathways. Diabetes 2010; 59: 3229–3239.
Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010; 49: 884–897.
Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes with a major role for the HLA. PLoS Genet 2011; 7: e1002197.
Cross-Disorder Group of the Psychiatric Genomics Consortium; Genetic Risk Outcome of Psychosis (GROUP) Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379.
Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium Ripke S, Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium Wray NR, Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium Lewis CM, Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium Hamilton SP, Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium Weissman MM, Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium Breen G et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013; 18: 497–511.
Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011; 43: 969–976.
Ruiz-Llorente S, Carrillo Santa de Pau E, Sastre-Perona A, Montero-Conde C, Gómez-López G, Fagin JA et al. Genome-wide analysis of Pax8 binding provides new insights into thyroid functions. BMC Genomics 2012; 13: 147.
Spiegel K, Leproult R, Van Cauter E . Impact of sleep debt on metabolic and endocrine function. Lancet 1999; 354: 1435–1438.
Kessler L, Nedeltcheva A, Imperial J, Penev PD . Changes in serum TSH and free T4 during human sleep restriction. Sleep 2010; 33: 1115–1118.
Krishnan PV, Vadivu AS, Alappatt A, Kameswaran M . Prevalence of sleep abnormalities and their association among hypothyroid patients in an Indian population. Sleep Med 2012; 13: 1232–1237.
Kales A, Heuser G, Jacobson A, Kales JD, Hanley J, Zweizig JR et al. All night sleep studies in hypothyroid patients before and after treatment. J Clin Endocrinol Metab 1967; 27: 1593–1599.
Marinò M, Chiovato L, Pinchera A . Graves’ disease. In Jameson JL, De Groot LJ (eds) Endocrinology. Saunders Elsevier: Philadelphia, PA, USA, pp 1527–1558 2010.
Opp MR, Krueger JM . Interleukin 1-receptor antagonist blocks interleukin 1-induced sleep and fever. Am J Physiol 1991; 260: R453–R457.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010; 42: 937–948.
Van den Berg JF, Van Rooij FJ, Vos H, Tulen JH, Hofman A, Miedema HM et al. Disagreement between subjective and actigraphic measures of sleep duration in a population-based study of elderly persons. J Sleep Res 2008; 17: 295–302.
Gerhman P, Seelig AD, Jacobson IG, Boykp EJ, Hooper TI, Gackstetter GD et al. Predeployment sleep duration and insomnia as risk factors for new-onset mental health disorders following military deployment. Sleep 2013; 36: 1009–1018.
Acknowledgements
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC–55022, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. The Cardiovascular Health Study was supported by NHLBI contracts HHSN268201200036C, N01-HC-85239, N01-HC-55222, N01-HC-85079, N01-HC-85080, N01-HC-85081, N01-HC-85082, N01-HC-85083, N01-HC-85086; and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institute on Aging (NIA). DNA handling and genotyping was supported in part by the National Center for Research Resources grant UL1RR033176, now at the National Center for Advancing Translational Sciences CTSI grant UL1TR000124; the National Institute of Diabetes and Digestive and Kidney Disease grant DK063491 to the Southern California Diabetes Endocrinology Research Center. This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (contract no. N01‐HC‐25195) and its contract with Affymetrix, Inc for genotyping services (contract no. N02‐HL‐6‐4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA‐II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The Health, Aging and Body Composition Study supported by NIA contracts N01-AG-62101, N01-AG-62103 and N01-AG-62106 and NIA grants 1R01AG032098-01A1 and 1R01AG030474-01A1. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, NIA. The Helsinki Birth Cohort Study has been supported by grants from the Academy of Finland, the Finnish Diabetes Research Society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Signe and Ane Gyllenberg Foundation, University of Helsinki, Ministry of Education, Ahokas Foundation, Emil Aaltonen Foundation, Juho Vainio Foundation, and Wellcome Trust (grant number WT089062). The Nurses Health Study and Health Professional Follow-Up Study GWAS were supported by grants from the National Institutes of Health [NCI (CA40356, CA087969, CA055075, CA98233), NIDDK (DK058845, DK070756), NHGRI (HG004399), NHLBI (HL35464)] with additional support from Merck/Rosetta Research Laboratories, North Wales, PA. The Invecchiare in CHIANTI study baseline (1998–2000) was supported as a ‘targeted project’ (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336). The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding. The following institutes provide support: the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Institute on Aging (NIA), the National Center for Research Resources (NCRR), and NIH Roadmap for Medical Research under the following grant numbers: U01 AR45580, U01 AR45614, U01 AR45632, U01 AR45647, U01 AR45654, U01 AR45583, U01 AG18197, U01-AG027810, and UL1 RR024140. The National Heart, Lung, and Blood Institute (NHLBI) provides funding for the MrOS Sleep ancillary study ‘Outcomes of Sleep Disorders in Older Men’ under the following grant numbers: R01 HL071194, R01 HL070848, R01 HL070847, R01 HL070842, R01 HL070841, R01 HL070837, R01 HL070838, and R01 HL070839’. The NIAMS provides funding for the MrOS ancillary study ‘GWAS in MrOS and SOF’ under the grant number RC2ARO58973. The Quebec Family Study was funded by multiple grants from the Medical Research Council of Canada and the Canadian Institutes for Health Research. This work was supported by a team grant from the Canadian Institutes for Health Research (FRN-CCT-83028). Funding for the Queensland Institute of Medical Research Twin Study was provided by the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498), the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016, DP0343921), the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254) and the US National Institutes of Health (NIH grants AA07535, AA10249, AA11998, AA13320, AA13321, AA13326, AA14041, MH66206). A portion of the genotyping on which this study was based (Illumina 370K scans) was carried out at the Center for Inherited Disease Research, Baltimore (CIDR), through an access award to our late colleague Dr Richard Todd (Psychiatry, Washington University School of Medicine, St Louis). Statistical analyses were carried out on the Genetic Cluster Computer, which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003). EMB is supported by NHMRC grant 613608. The generation and management of GWAS genotype data for the Rotterdam Study are supported by the Netherlands Organisation of Scientific Research NWO Investments (no. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810. The Rotterdam Study is funded by Erasmus MC and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. Henning Tiemeier was supported by the VIDI grant of ZonMw (2009-017.106.370). Karin Hek was supported by a grant from BavoEuropoort. Netherlands Twin Registry funding was obtained from the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW grants 904-61-090, 985-10-002, 904-61-193,480-04-004, 400-05-717, Addiction-31160008, Middelgroot-911-09-032, Spinozapremie 56-464-14192), Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI –NL, 184.021.007), VU University’s Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA), European Science Foundation (ESF, EU/QLRT-2001-01254), the European Community’s Seventh Framework Program (FP7/2007-2013), ENGAGE (HEALTH-F4-2007-201413); European Science Council (ERC 230374), Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls, South Dakota (USA), and the National Institutes of Health (NIH, R01D0042157-01A, Grand Opportunity grants 1RC2MH089951-01 and 1RC2 MH089995-01). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. The Study of Health in Pomerania is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103 and 01ZZ0403), the Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens AG and the Caché Campus program of the InterSystems GmbH. The Study of Osteoporotic Fractures is supported by National Institutes of Health funding. The National Institute on Aging (NIA) provides support under the following grant numbers: R01 AG005407, R01 AR35582, R01 AR35583, R01 AR35584, R01 AG005394, R01 AG027574, R01 AG027576, and R01 AG026720. The National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) provides funding for the ancillary study ‘GWAS in MrOS and SOF’ under the grant number RC2ARO58973. TwinsUK was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013), ENGAGE project grant agreement (HEALTH-F4-2007-201413). The study also receives support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London. Genotyping was performed by the Wellcome Trust Sanger Institute, support of the National Eye Institute via an NIH/CIDR genotyping project. This research was supported for the Wisconsin Sleep Cohort Study by the National Heart, Lung, and Blood Institute (R01HL62252) and National Center for Research Resources (1UL1RR025011) and by NS23724. The Young Finns Study has been financially supported by the Academy of Finland: grants 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi), the Social Insurance Institution of Finland, Kuopio, Tampere and Turku University Hospital Medical Funds (grant 9M048 for 9N035 for TeLeht), Juho Vainio Foundation, Paavo Nurmi Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation, Tampere Tuberculosis Foundation and Emil Aaltonen Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Rights and permissions
About this article
Cite this article
Gottlieb, D., Hek, K., Chen, Th. et al. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol Psychiatry 20, 1232–1239 (2015). https://doi.org/10.1038/mp.2014.133
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2014.133
This article is cited by
-
Combining MTI-31 with RAD001 inhibits tumor growth and invasion of kidney cancer by activating autophagy
Journal of Applied Genetics (2024)
-
Genetics of circadian rhythms and sleep in human health and disease
Nature Reviews Genetics (2023)
-
Genome-wide association studies and cross-population meta-analyses investigating short and long sleep duration
Nature Communications (2023)
-
Association of physical activity, sedentary behaviours and sleep duration with cardiovascular diseases and lipid profiles: a Mendelian randomization analysis
Lipids in Health and Disease (2020)
-
The role of the circadian system in the etiology and pathophysiology of ADHD: time to redefine ADHD?
ADHD Attention Deficit and Hyperactivity Disorders (2019)