Abstract
The appendix gives rise to goblet cell carcinoids, which represent special carcinomas with distinct biological and histological features. Their genetic background and molecular relationship to colorectal adenocarcinoma is largely unknown. We therefore performed a next-generation sequencing analysis of 25 appendiceal carcinomas including 11 goblet cell carcinoids, 7 adenocarcinomas ex-goblet cell carcinoid, and 7 primary colorectal-type adenocarcinomas, using a modified Colorectal Cancer specific Panel comprising 32 genes linked to colorectal and neuroendocrine tumorigenesis. The mutational profiles of these neoplasms were compared with those of conventional adenocarcinomas, mixed adenoneuroendocrine carcinomas, and neuroendocrine carcinomas of the colorectum. In addition, a large-scale pan-cancer sequencing panel covering 409 genes was applied to selected cases of goblet cell carcinoid/adenocarcinoma ex-goblet cell carcinoid (n=2, respectively). Mutations in colorectal cancer-related genes (eg, TP53, KRAS, APC) were rare to absent in both, goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid, but frequent in primary colorectal-type adenocarcinomas of the appendix. Additional large-scale sequencing of selected goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid revealed mutations in Wnt-signaling-associated genes (USP9X, NOTCH1, CTNNA1, CTNNB1, TRRAP). These data suggest that appendiceal goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid constitute a morphomolecular entity, histologically and genetically distinct from appendiceal colorectal-type adenocarcinomas and its colorectal counterparts. Altered Wnt-signaling associated genes, apart from APC, may act as potential drivers of these neoplasms. The absence of KRAS/NRAS mutations might render some of these tumors eligible for anti-EGFR directed therapy regimens.
Similar content being viewed by others
Main
The colorectal-type epithelium of the appendiceal mucosa1 gives rise to a variety of comparatively rare and histologically heterogeneous mucus producing carcinomas.2, 3, 4, 5 Their spectrum ranges from primary colorectal-type adenocarcinoma, which is morphologically similar to its counterpart in the large intestine,4, 5 to goblet cell carcinoid, which represents a distinctive appendiceal carcinoma with glands showing inconspicuous lumina, which are composed of mucus producing cells resembling goblet cells and scattered neuroendocrine cells, most of which produce serotonin.6, 7 Between these two extremes of the spectrum are low-grade mucinous neoplasms and a group of neoplasms called 'adenocarcinomas ex-goblet cell carcinoid', a term that has initially been proposed by Tang et al4 for a subgroup of clinically aggressive goblet cell carcinoids.7, 8, 9 The latter tumors show a variety of morphologically distinct, adenocarcinoma-like growth patterns (eg, microglandular, solid sheet-like) adjacent to areas resembling conventional goblet cell carcinoid.4, 6, 7 In the current WHO classification, these neoplasms are listed among mixed adenoneuroendocrine carcinomas of the appendix.5
Although the genetic profiles of colorectal adenocarcinomas,10, 11, 12 colorectal mixed adenoneuroendocrine carcinomas, and neuroendocrine carcinomas13, 14 have been well described, little is known about the molecular alterations underlying appendiceal carcinomas and their genetic relationship towards colorectal carcinomas. Although one study described high frequencies of KRAS/ GNAS mutations in low-grade appendiceal mucinous neoplasms15 and postulated frequent TP53 alterations in a subgroup of their investigated cohort referred to as 'appendiceal cancers',15 no comprehensive next-generation sequencing based study of goblet cell carcinoids, adenocarcinomas ex-goblet cell carcinoid and primary colorectal-type adenocarcinomas of the appendix has been performed so far.
In this study, we examined the genetic profile of appendiceal carcinomas including 11 goblet cell carcinoids, 7 adenocarcinomas ex-goblet cell carcinoid, and 7 primary colorectal-type adenocarcinomas of the appendix, applying a modified Colorectal Cancer Panel covering 32 genes linked to colorectal and neuroendocrine tumorigenesis and compared their mutational profiles with those of colorectal adenocarcinomas, colorectal mixed adenoneuroendocrine carcinomas, and colorectal neuroendocrine carcinomas.10, 13 Selected cases of goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid (n=2, respectively) were investigated using the Comprehensive Cancer Panel, which covers most of the exonic regions of 409 cancer-related genes.
The aim of this study was to compare the genetic profile of appendiceal goblet cell carcinoids with those of adenocarcinomas ex-goblet cell carcinoid and primary colorectal-type adenocarcinomas of the appendix, and finally, with those of colorectal adenocarcinomas as well as colorectal mixed adenoneuroendocrine carcinomas and neuroendocrine carcinomas. Moreover, we searched for genetic alterations representing potential genetic drivers of these neoplasms. Finally, we screened their mutational profiles for drugable alterations.
Materials and methods
Case Selection
Paraffin-embedded, formalin-fixed tissues of resected tumors of the appendix were retrieved from the archives and consultation files of the Institute of Pathology at the Technical University of Munich (Munich, Germany) and from the archives of the Institute of Pathology of the University Hospital of Mainz (Mainz, Germany), and three subgroups of appendiceal carcinomas were analyzed: goblet cell carcinoids (11 cases), adenocarcinomas that show various growth patterns and appear to arise from goblet cell carcinoids ('adenocarcinomas ex-goblet cell carcinoid'; seven cases) and primary colorectal-type adenocarcinomas of the appendix (seven cases).5, 6, 7 Mucinous adenocarcinomas, low-grade appendiceal mucinous neoplasms, and pure neuroendocrine neoplasms were excluded.5
Clinicopathological Data
Clinical, demographic, and macroscopic data were obtained by review of the medical charts and pathology reports. Pathological staging followed the UICC TNM staging system for carcinomas of the appendix.2
Histological Analyses
Hematoxylin and eosin, and Periodic acid Schiff stained sections were reviewed by three pathologists (MJ, GK, and AMS) and histologically classified in accordance to the current WHO classification of tumors of the digestive system5 and under consideration of the published criteria for the diagnosis of 'adenocarcinoma ex-goblet cell carcinoid' by Tang et al and Reid et al.6, 7 In addition, all tumors were investigated for the presence/absence of associated non-invasive adenomatous precursor lesions.5
Immunohistochemical Analyses
Three micrometer-thick paraffin sections from goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid were immunohistochemically stained for synaptophysin (Ventana Medical Systems, Tuscon, USA; 1:1), Ki-67 (DakoCytomation, Glostrup, Denmark; 1:50), serotonin (PROGEN Biotechnik GmbH, Heidelberg, Germany; 1:60), MLH1 (Ventana Medical Systems; 1:1), PMS2 (Ventana Medical Systems; 1:1), MSH2 (Menarini Diagnostics, Florence, Italy; 1:75), MSH6 (BD Biosciences, Franklin Lakes, USA, 1:400), and CDX2 (DakoCytomation; 1:200), using a fully automated slide preparation system (‘Benchmark XT System’; Ventana Medical Systems). The expression of synaptophysin, serotonin und CDX2 (nuclear expression) was scored according to the percentage of positive cells: negative, focally positive (1–50%) and diffusely positive (>50%). Ki-67-positive cells were counted in two representative high power fields including hotspot areas, and at least 500 tumor cells per case.
DNA Preparation
Tumor areas were marked on a hematoxylin and eosin stained slide, and the respective tumor cell content was estimated by two pathologists (MJ, AMS) and documented for each case. Cases with a tumor cell content of >20% per tissue sample were considered appropriate for genetic analysis and the respective tumor tissue was manually microdissected from subsequent unstained slides for DNA extraction. Genomic DNA was extracted using the Maxwell 16 Research System (Promega, Madison, USA) after proteinase K digestion by fully automated purification. DNA content was measured fluorimetrically using the QuBit HS DNA Assay (Thermo Fisher Scientific, Waltham, USA). Real-time qPCR-based method (RNAseP Detection system, Thermo Fisher Scientific) was utilized to confirm sequencing grade quality.16, 17
Colorectal Cancer Panel
All cases were investigated with our custom-designed Colorectal Cancer Panel. As described previously, the multiplex PCR-based Ion Torrent AmpliSeq technology (Thermo Fisher Scientific) combined with our custom-designed Colorectal Cancer Panel, to which 16 amplicons covering hotspot regions from RB1 and RET were added, were used for library preparation.10, 18, 19 In total, our modified Colorectal Cancer Panel consists of one primer pool yielding 196 amplicons covering hotspot regions of 32 genes and the Ion AmpliSeq Library Kit v2.0 was used for library preparation.13 Details regarding our customized Colorectal Cancer Panel can be seen in Supplementary Table 1. Library preparation was performed according to the manufacturers protocol (Thermo Fisher Scientific), as described previously.10, 18, 19 Approximately 10 ng of DNA, determined by qPCR assay, were used for amplification. After dilution of the individual libraries to a final concentration of 25 pM, 10 of these libraries were pooled. Using the Ion 520/530 Chef Kit on an Ion Chef instrument, the libraries were processed for sequencing utilizing the Ion S5 Sequencing chemistry and loaded onto a 520 chip. Sequencing was performed using an Ion S5XL instrument (Thermo Fisher Scientific).
Comprehensive Cancer Panel
In four selected cases (two goblet cell carcinoids, two adenocarcinomas ex-goblet cell carcinoid) additional massive parallel sequencing was performed using the AmpliSeq Comprehensive Cancer Panel (AmpliSeq CCP, Thermo Fisher Scientific), which nearly covers the complete exonic regions of 409 cancer-related genes and consists of 15 992 amplicons spread out over four primer pools. Library preparation and sequencing was performed as previously described,20 a list of all genes included in this panel is given in Supplementary table 2.
Data Analysis
Alignment against the human genome (version hg19) was performed using the Torrent Mapping Alignment Program algorithm and the implemented Torrent Suite Software (version 5.2.2.) was used for processing of raw sequencing data. For DNA mutation analysis the build-in Variant Caller plugin was used. Variant annotation was performed using a custom built variant annotation pipeline based on ANNOVAR21 and sequencing reads were visualized using the Integrative Genomics Viewer Browser (IGV, http://www.broadinstitute.org/igv/). For the identification of already known somatic mutations and the determination of mutation types, variants were screened against the Catalogue of Somatic Mutations in Cancer database.22 The Single Nucleotide Polymorphism database23 and the Exome Variant Server (http://evs.gs.washington.edu/EVS/) were screened to exclude common germline variants from the analysis. Non-synonymous variants with an allele frequency ≥5% were reported as mutations.
Prediction of Copy Number Variations
The evaluation of copy number variations (amplifications and deletions) was performed using the coverage data obtained from the Torrent Suite software through analysis of the coverage data summary for each sample and each amplicon, as described previously.17, 24
Sequencing Performance
A mean number of 696 185 reads (range: 356 538–1 084 544) was achieved using the modified Colorectal Cancer panel for targeted sequencing and a mean number of reads per amplicon of 3589 (range: 169–11 942) was reached. Two amplicons of the modified Colorectal Cancer Panel showed a mean coverage of <20 reads and were excluded from the CNV analysis. Mean numbers of reads of the AmpliSeq Comprehensive Cancer panel was 9 413 454 (range: 8 240 857–10 945 795), mean number of reads per amplicon was 589 (range: 20–12 780). Of the AmpliSeq Comprehensive Cancer Panel, 331 amplicons revealed a mean coverage of <20 reads and were excluded from CNV analysis.
Microsatellite Instability Analysis
Microsatellite instability analysis was performed dually. First, all tumors were immunohistochemically investigated using antibodies against the DNA-mismatch repair proteins MLH1, MSH2, MSH6, and PMS2.25 Loss of nuclear staining in >90% of tumor cells was reported as microsatellite instable. Furthermore, the marker panel BAT25, BAT26, DSS346, D2S123, and DI7S250 was applied as described previously. High microsatellite instability was scored if at least two of five markers showed genetic instability.26
Colorectal Adenocarcinoma Control Group
The genetic data of the appendiceal carcinomas were compared with those from our large, previously published data set of 196 colorectal adenocarcinomas, which were genetically investigated using a similar custom-designed Colorectal Cancer Panel.10 In addition, the genetic data of the appendiceal carcinomas were compared with those from a previously published data set of 19 colorectal mixed adenoneuroendocrine carcinomas and 8 colorectal neuroendocrine carcinomas (seven large cell type, one small cell type; no RB1 mutations), that were investigated using our modified Colorectal Cancer Panel.13
Statistical Analyses
Statistical analyses were performed using the Statistic Package for Social Sciences 23.0 statistical software (SPSS, Chicago, USA). Correlation analyses between genetic data and clinicopathological parameters were performed using Fisher’s exact test. A P-value <0.05 was considered as significant.
The study has been approved by the local ethics committee of the Technical University of Munich (reference number: 503/16s).
Results
Clinicopathological Features
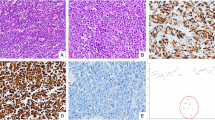
The male to female ratio of the 11 patients with goblet cell carcinoids was 6/5, and the mean age at time of diagnosis was 61 years (Table 1). Of these patients, 73% (8/11) had symptoms suggestive of appendicitis. All tumors showed advanced stages (pT3: 9/11, 82%; pT4: 2/11, 18%), but no nodal involvement. Only one case showed distant metastases (1/11, 9%; uterus). Histologically, all goblet cell carcinoids were uniformly composed of goblet cells resembling crypt cells, which formed glandular tumor cell clusters lacking distinct lumina (Figure 1). Well-differentiated adenocarcinoma-like glands, poorly cohesive growth patterns, or high-grade cytomorphologic features were not observed in these cases.6
Histomorphology of appendiceal carcinomas: (a+b): Conventional goblet cell carcinoids, with synaptophysin positivity (b inset), (c) Adenocarcinoma ex-goblet cell carcinoid with adenocarcinoma-like glands (star) adjacent to areas resembling conventional goblet cell carcinoid (arrow), (d) Adenocarcinoma ex-goblet cell carcinoid displaying a solid sheet-like growth pattern next to areas with a conventional goblet cell carcinoid morphology (d inset), (e) High magnification of primary colorectal-type adenocarcinoma of the appendix, (f) Villous adenoma of the appendix with low-grade intraepithelial neoplasia and adjacent invasive colorectal-type adenocarcinoma (f inset).
Seven patients were diagnosed with adenocarcinoma ex-goblet cell carcinoid, and of those, four were men and three were women. Mean patient age in this subgroup was 55.5 years. All tumors with known pT-status were in an advanced local stage and some already showed serosal penetration (pT3: 2/7, 28%; pT4: 3/7, 44%; pTx: 2/7, 28%). One case (1/7; 14%) showed metastatic spread to local lymph nodes, three cases had distant metastases (3/8, 38%; liver, uterus, peritoneum). Histologically, all tumors contained small, but distinctive areas representing conventional goblet cell carcinoid, which alternated with other distinct growth patterns, as defined by Reid and colleagues6 (eg, poorly cohesive goblet cell pattern, goblet cell carcinoid pattern with high-grade cytomorphology, or solid sheet-like growth pattern) (Figure 1). Taken together, the various adenocarcinoma patterns were the predominant tumor component in all cases of adenocarcinoma ex-goblet cell carcinoid. An overview of all identifiable growth patterns per adenocarcinoma ex-goblet cell carcinoid, including their percentage within each case, is provided in Supplementary Table 3.
Seven patients in our tumor series were diagnosed with primary colorectal-type adenocarcinoma of the appendix (men: women, 4/3; mean age: 71 years). Most of these tumors were in an advanced local stage (pT3/4: 6/7, 86%), whereas in one case, the infiltration was limited to submucosal tissue (pT1: 1/7, 14%). All colorectal-type adenocarcinomas were of conventional type, special histologic variants (eg, medullary carcinoma, mucinous carcinoma) were not present. As precursors to these tumors, adenomas were identified in 5/7 cases (tubulovillous adenoma: n=2, villous adenoma: n=2, tubular adenoma: n=1). Only one case showed a lymph node metastasis (1/7, 14%), distant metastases were absent.
Immunohistochemical Analysis
All goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid showed immunostaining for synaptophysin at least in a fraction of tumor cells, ranging from focal to almost diffuse. Serotonin was expressed in four goblet cell carcinoids and in three adenocarcinomas ex-goblet cell carcinoids. CDX2 was diffusely expressed in all goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid, underlining the hypothesis that these tumors derive from a colorectal-type epithelium (Figure 1 and Table 1). Scattered synaptophysin-positive cells (max. 3% of tumor cells) were found in 2/7 primary colorectal-type adenocarcinomas of the appendix.
Microsatellite Instability Analysis
Immunohistochemical microsatellite instability analysis of all goblet cell carcinoids, adenocarcinomas ex-goblet cell carcinoid and primary colorectal-type adenocarcinomas of the appendix revealed a preserved nuclear expression of MLH1, MSH2, MSH6, and PMS2 in all tumors. Consistent with the immunohistochemical data, none of the goblet cell neoplasms or primary colorectal-type adenocarcinomas showed signs of microsatellite instability in the additional molecular microsatellite instability analysis. Supplementary Table 5 gives an overview of the results of microsatellite instability testing for each individual neoplasm.
Genetic Profiling of Goblet Cell Carcinoids
As depicted in Figure 2 and Supplementary Table 4, 4/11 goblet cell carcinoids showed a total of four singular mutations in 4/32 genes included in our Colorectal Cancer Panel. One goblet cell carcinoid (GCC01) showed a TP53 mutation, another case (GCC10) harbored a non V600E BRAF mutation. One tumor (GCC09) displayed a singular TGFBR2 mutation, whereas another neoplasm (GCC03) had an ERBB2 mutation. Alterations in other major colorectal adenocarcinoma-related genes such as APC, KRAS, NRAS, PIK3CA, PTEN, or SMAD4 were not detected (Figure 2). Copy number variations were not detected.
Distribution of colorectal adenocarcinoma-related mutations in appendiceal carcinomas: primary colorectal-type adenocarcinomas of the appendix show frequent alterations in genes classically linked to colorectal carcinogenesis, which are rather scarce in both conventional goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid. Each colored box represents mutations identified in the samples. Color annotations: orange=missense mutation; purple=nonsense mutation; green=splice site variant; blue=frameshift; red=delins. Abbreviations: GCC=Goblet cell carcinoid; AdenoexGCC=Adenocarcinoma ex-goblet cell carcinoid; AppCRC=Primary colorectal-type adenocarcinoma of the appendix.
Genetic Profiling of Adenocarcinomas Ex-goblet Cell Carcinoid
Initial massive parallel sequencing revealed a total of three mutations in 3/32 genes of our Colorectal Cancer Panel. In all adenocarcinomas ex-goblet cell carcinoid, the entire tumor tissue (including every identifiable adenocarcinoma component and areas resembling conventional goblet cell carcinoid), was genetically investigated. One adenocarcinoma ex-goblet cell carcinoid (AdenoexGCC05) showed a non V600E BRAF mutation, whereas another carcinoma (AdenoexGCC07) harbored a SMAD4 mutation and another neoplasm showed a CTNNB1 mutation (AdenoexGCC04) (Figure 2, Supplementary Table 4). Copy number variations were not detected.
Genetic Profiling of Primary Colorectal-type Adenocarcinomas of the Appendix
Fourteen somatic mutations in seven colorectal adenocarcinoma-related genes were identifiable in 6/7 primary colorectal-type adenocarcinomas of the appendix. Alterations of TP53 (5/7) and KRAS (3/7) were most frequent, two carcinomas harbored PIK3CA mutations (2/7). APC (1/7), BRAF (1/7), ATM (1/7), and CTNNB1 (1/7) were each mutated in one case (Figure 2; Supplementary Table 4). Copy number variations were not detected.
Comparison of the Mutational Profiles
Goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid did not differ in their mutational profiles. KRAS (P=0.036) was more frequently altered in appendiceal colorectal-type adenocarcinomas than in goblet cell carcinoids. TP53 mutations were more common in appendiceal colorectal-type adenocarcinomas than in goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid (P=0.01).
As depicted in Figure 4, no differences between the mutational profiles of primary colorectal-type adenocarcinomas of the appendix and those of conventional colorectal carcinomas, colorectal mixed adenoneuroendocrine carcinomas, and colorectal neuroendocrine carcinomas were detected.
Goblet cell carcinoids and colorectal adenocarcinomas significantly differed in their mutational profiles, as TP53- (P<0.001), KRAS- (P<0.001), and APC- (P<0.001) mutations were frequent in conventional colorectal adenocarcinomas, but rare to absent in goblet cell carcinoids. TP53- (P=0.018) and APC (P=0.04) mutations were also more common in colorectal neuroendocrine carcinomas. Colorectal mixed adenoneuroendocrine carcinomas were more frequently TP53 mutated (P=0.027) than goblet cell carcinoids.
The frequency of TP53 (P<0.001), KRAS (P<0.02), and APC (P<0.001) mutations in adenocarcinomas ex-goblet cell carcinoid was significantly lower than in colorectal adenocarcinomas. Furthermore, TP53 mutations were less frequent in adenocarcinomas ex-goblet cell carcinoid than in colorectal mixed adenoneuroendocrine carcinomas (P=0.02) and colorectal neuroendocrine carcinomas (P=0.013), which were additionally more often APC mutated (P=0.013).
Additional Large-Scale Sequencing of Goblet Cell Carcinoids and Adenocarcinomas Ex-goblet Cell Carcinoid
Additional large-scale sequencing (Comprehensive Cancer Panel, 409 genes; 15992 amplicons) of two conventional goblet cell carcinoids revealed a singular USP9X mutation in one tumor, for which a TP53 mutation was detected by our initial sequencing approach (GCC01) and coexisting NOTCH1, NUMA1, and CTNNA1 mutations in another case (GCC04) (Figure 3, Supplementary Table 4).
Comparison of the genetic background of the diverse appendiceal carcinomas and colorectal adenocarcinoma. Hexagons show genes that are either classically colorectal adenocarcinoma-related or have at least been substantially associated with colorectal cancer, circles indicate mutations detected in goblet cell carcinoid/adenocarcinoma ex-goblet cell carcinoid, which are not classically associated with colorectal carcinogenesis.
Additional large-scale sequencing (Comprehensive Cancer Panel) of two adenocarcinomas ex-goblet cell carcinoid revealed a total of five mutations in one case (AdenoexGCC04: CTNNB1, LIFR, TRRAP, KMT2D, TAF1), whereas the other carcinoma displayed a FGFR1 stop-gain mutation (AdenoexGCC06) (Figure 3; Supplementary Table 4).
Worthy of note, three of the four goblet cell related neoplasms, that underwent additional large-scale sequencing, harbored mutations in genes (USP9X, NOTCH1, CTNNA1, CTNNB1, TRRAP), which are known to be directly involved or to interact with the Wnt-signaling pathway.27, 28, 29, 30, 31, 32
Discussion
We investigated the mutational profiles of 25 appendiceal goblet cell carcinoids, adenocarcinomas ex-goblet cell carcinoid, and primary colorectal-type adenocarcinomas via targeted next-generation sequencing and compared the results with previously published data from a large colorectal adenocarcinoma cohort (196 patients)10 and a series of 27 patients with either colorectal mixed adenoneuroendocrine carcinoma or neuroendocrine carcinoma.13 As illustrated in Figures 2,3, and 4, our genetic analysis revealed, that the mutations in primary colorectal-type adenocarcinomas of the appendix are similar to those in colorectal adenocarcinomas, indicating that the phenotypic relationship of these neoplasms is based on a common genetic profile, which is characterized by shared alterations in colorectal adenocarcinoma-related genes such as TP53/KRAS/BRAF/PIK3CA.
Comprehensive analysis of the mutational profiles of colorectal adenocarcinoma, colorectal mixed adenoneuroendocrine carcinoma, and neuroendocrine carcinoma with those of primary colorectal-type adenocarcinoma of the appendix and conventional goblet cell carcinoid as well as adenocarcinoma ex-goblet cell carcinoid. Note the rather uniform distribution of colorectal adenocarcinoma-related mutations between colorectal neoplasms and primary colorectal-type adenocarcinoma of the appendix. Color annotations: gray=Colorectal adenocarcinoma; light blue=Colorectal mixed adenoneuroendocrine carcinoma; turquoise=Colorectal neuroendocrine carcinoma; black=Primary colorectal-type adenocarcinoma of the appendix; red=Goblet cell carcinoid; orange=Adenocarcinoma ex-goblet cell carcinoid. Abbreviations: GCC=Conventional goblet cell carcinoid; AdenoexGCC=Adenocarcinoma ex-goblet cell carcinoid; AppCRC=Primary colorectal-type adenocarcinoma of the appendix; CRC=Colorectal adenocarcinoma; MANEC=Colorectal mixed adenoneuroendocrine carcinoma; NEC=Colorectal neuroendocrine carcinoma.
The same alterations, however, were rare to absent in both, goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid. These data indicate that the latter two tumor groups appear to be genetically distinct from the colorectal carcinoma subgroup. Moreover, they suggest that adenocarcinomas ex-goblet cell carcinoid indeed derive from goblet cell carcinoids. Although we identified isolated colorectal adenocarcinoma-related mutations10, 11 in few goblet cell carcinoids (4/11 cases) and adenocarcinomas ex-goblet cell carcinoid (3/7 cases), they probably have no significant role as genetic drivers in these neoplasms, as they were not repeatedly detected and found to be scattered throughout the respective cohorts. Nevertheless, we cannot exclude with absolute certainty, that less-common colorectal adenocarcinoma-related gene mutations, which were not present in our investigated cases, might be of some importance for the tumorigenesis of these neoplasms, as our tumor series of goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoids was rather small (n=18). Furthermore, although goblet cell neoplasms and neuroendocrine tumors of the appendix or the small intestine ('pure carcinoids') show differences concerning their morphology5 and their clinical course,33 our study is not able to make a clear statement regarding the genetic relationship between these entities, as CDKN1B, the only gene that was found to be recurrently altered in a subset of (small intestinal) neuroendocrine tumors,34 was not included in the applied sequencing panels. Therefore, the significance of CDKN1B for the development of goblet cell neoplasms of the appendix should be the subject of further studies.
The finding, that the genetic similarities between adenocarcinomas ex-goblet cell carcinoid and colorectal mixed adenoneuroendocrine carcinomas appear to be quite limited,13 is particularly remarkable, as it suggests that the two tumor groups do not share common carcinogenic pathways, although both arise from a colorectal-type mucosa and show an admixture of adenocarcinoma components with neuroendocrine differentiated tumor cells. Our findings are at variance with genetic data from a previous next-generation sequencing based study by Liu and colleagues.15 They investigated 'appendiceal adenocarcinomas with goblet cell features' and reported frequent TP53, SMAD4, APC, and PIK3CA mutations, which were rare to absent in all goblet cell carcinoid related neoplasms included in our tumor series (including cases with additional expanded sequencing), but comparatively frequent within primary colorectal-type adenocarcinomas of the appendix. Although we do not have a ready explanation for this discrepancy, it might be that Liu´s cohort of appendiceal adenocarcinomas was histopathologically heterogeneous and contained some colorectal-type adenocarcinomas.
As the first round of genetic testing showed only few colorectal adenocarcinoma-related mutations in goblet cell carcinoids and their ex-goblet cell carcinoid variants, we decided to take our sequencing approach one step further and investigated selected cases from both subgroups with a large-scale next-generation sequencing panel, which covers nearly all exons from 409 cancer-related genes. Interestingly, both goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid harbored alterations in genes (USP9X, NOTCH1, CTNNA1, and CTNNB1, TRRAP, respectively), whose transcriptional products are known to be directly involved or interact with the Wnt-signaling pathway,27, 28, 29, 30, 31, 32 that is thought to have an important role in intestinal proliferation and differentiation.35, 36 It is therefore possible, that alterations of Wnt-signaling pathway-associated proteins, beside APC, may be of significance for the development of appendiceal goblet cell carcinoids and their high-grade variants. As the hypothesis of an APC-independent involvement of the Wnt-signaling pathway is only based on the analysis of a few cases, it needs to be confirmed in more extended studies.
Consistent with data from previous studies,37, 38 KRAS/NRAS mutations as well as microsatellite instability were lacking in the goblet cell carcinoid group. Considering the rarity of somatic mutations in genes (BRAF, PIK3CA)39, 40, 41 related to anti-EGFR resistance in RASwt tumors, our findings might indicate a potential effectiveness of anti-EGFR directed therapy regimens in these entities.
In conclusion, our broad spectrum next-generation sequencing analysis of 25 appendiceal carcinomas established a close morphogenetic relationship of primary colorectal-type adenocarcinoma of the appendix with conventional colorectal adenocarcinoma. In addition, the data demonstrated that appendiceal goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid have a common genetic basis and constitute a morphomolecular entity of the appendix, whose tumorigenesis appears to differ not only from that of colorectal carcinomas, but also from that of colorectal mixed adenoneuroendocrine carcinomas and neuroendocrine carcinomas.
References
Katzin WE, Petras RE Veriform appendix. In: Mills SE For Pathologists. Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2012, pp 698–704.
Brierley JD, Gospodarowicz MK, Wittekind C Appendix. In: TNM classification of malignant tumours 8th edn. John Wiley & Sons: Hoboken, NJ, USA, 2017, pp 93–96.
Carr NJ, McCarthy WF, Sobin LH . Epithelial noncarcinoid tumors and tumor-like lesions of the appendix. A clinicopathologic study of 184 patients with a multivariate analysis of prognostic factors. Cancer 1995;75:757–768.
Tang LH . Epithelial neoplasms of the appendix. Arch Pathol Lab Med 2010;134:1612–1620.
Carr NJ, Sobin LH Adenocarcinoma of the appendix. In: Bosman FT, Carneiro F, Hruban RH, Theise ND WHO classification of tumours of the digestive system, World Health Organization2010,4th edn.WHO Press, IARC: Lyon, France, 2010, pp 122–129.
Reid MD, Basturk O, Shaib WL et al. Adenocarcinoma ex-goblet cell carcinoid (appendiceal-type crypt cell adenocarcinoma) is a morphologically distinct entity with highly aggressive behavior and frequent association with peritoneal/intra-abdominal dissemination: an analysis of 77 cases. Mod Pathol 2016;29:1243–1253.
Tang LH, Shia J, Soslow RA et al. Pathologic classification and clinical behavior of the spectrum of goblet cell carcinoid tumors of the appendix. Am J Surg Pathol 2008;32:1429–1443.
Hristov AC, Young RH, Vang R et al. Ovarian metastases of appendiceal tumors with goblet cell carcinoidlike and signet ring cell patterns: a report of 30 cases. Am J Surg Pathol 2007;31:1502–1511.
Taggart MW, Abraham SC, Overman MJ, Mansfield PF, Rashid A . Goblet cell carcinoid tumor, mixed goblet cell carcinoid-adenocarcinoma, and adenocarcinoma of the appendix: comparison of clinicopathologic features and prognosis. Arch Pathol Lab Med 2015;139:782–790.
Jesinghaus M, Pfarr N, Endris V et al. Genotyping of colorectal cancer for cancer precision medicine: results from the IPH center for molecular pathology. Genes Chromosomes Cancer 2016;55:505–521.
Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–337.
Guinney J, Dienstmann R, Wang X et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–1356.
Jesinghaus M, Konukiewitz B, Keller G et al. Colorectal mixed adenoneuroendocrine carcinomas and neuroendocrine carcinomas are genetically closely related to colorectal adenocarcinomas. Mod Pathol 2017;30:610–619.
Woischke C, Schaaf CW, Yang HM et al. In-depth mutational analyses of colorectal neuroendocrine carcinomas with adenoma or adenocarcinoma components. Mod Pathol 2016;30:95–103.
Liu X, Mody K, de Abreu FB et al. Molecular profiling of appendiceal epithelial tumors using massively parallel sequencing to identify somatic mutations. Clin Chem 2014;60:1004–1011.
Stenzinger A, Pfarr N, Penzel R et al. Semiconductor-based sequencing of formalin-fixed, paraffin-embedded colorectal cancer samples. Oncologist 2015;20:e10–e11.
Endris V, Penzel R, Warth A et al. Molecular diagnostic profiling of lung cancer specimens with a semiconductor-based massive parallel sequencing approach: feasibility, costs, and performance compared with conventional sequencing. J Mol Diagn 2013;15:765–775.
Jesinghaus M, Wolf T, Pfarr N et al. Distinctive spatiotemporal stability of somatic mutations in metastasized microsatellite-stable colorectal cancer. Am J Surg Pathol 2015;39:1140–1147.
Jesinghaus M, Pfarr N, Kloor M et al. Genetic heterogeneity in synchronous colorectal cancers impacts genotyping approaches and therapeutic strategies. Genes Chromosomes Cancer 2015;55:268–277.
Pfarr N, Darb-Esfahani S, Leichsenring J et al. Mutational profiles of Brenner tumors show distinctive features uncoupling urothelial carcinomas and ovarian carcinoma with transitional cell histology. Genes Chromosomes Cancer 2017;56:758–766.
Wang K, Li M, Hakonarson H . ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164.
Forbes SA, Beare D, Gunasekaran P et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015;43:D805–D811.
Sherry ST, Ward MH, Kholodov M et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–311.
Pfarr N, Penzel R, Klauschen F et al. Copy number changes of clinically actionable genes in melanoma, non-small cell lung cancer and colorectal cancer-A survey across 822 routine diagnostic cases. Genes Chromosomes Cancer 2016;55:821–833.
Lindor NM, Burgart LJ, Leontovich O et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 2002;20:1043–1048.
Boland CR, Goel A . Microsatellite instability in colorectal cancer. Gastroenterology 2010;138:2073–2087 e2073.
Foltz DR, Santiago MC, Berechid BE, Nye JS . Glycogen synthase kinase-3beta modulates notch signaling and stability. Curr Biol 2002;12:1006–1011.
Finkbeiner MG, Sawan C, Ouzounova M, Murr R, Herceg Z . HAT cofactor TRRAP mediates beta-catenin ubiquitination on the chromatin and the regulation of the canonical Wnt pathway. Cell Cycle 2008;7:3908–3914.
Su LK, Vogelstein B, Kinzler KW . Association of the APC tumor suppressor protein with catenins. Science 1993;262:1734–1737.
Taya S, Yamamoto T, Kanai-Azuma M, Wood SA, Kaibuchi K . The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells 1999;4:757–767.
Yang B, Zhang S, Wang Z et al. Deubiquitinase USP9X deubiquitinates beta-catenin and promotes high grade glioma cell growth. Oncotarget 2016;7:79515–79525.
Premarathne S, Murtaza M, Matigian N, Jolly LA, Wood SA . Loss of Usp9x disrupts cell adhesion, and components of the Wnt and Notch signaling pathways in neural progenitors. Sci Rep 2017;7:8109.
Rossi RE, Luong TV, Caplin ME et al. Goblet cell appendiceal tumors—management dilemmas and long-term outcomes. Surg Oncol 2015;24:47–53.
Francis JM, Kiezun A, Ramos AH et al. Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nat Genet 2013;45:1483–1486.
Clevers H, Nusse R . Wnt/beta-catenin signaling and disease. Cell 2012;149:1192–1205.
Krausova M, Korinek V . Wnt signaling in adult intestinal stem cells and cancer. Cell Signal 2014;26:570–579.
Dimmler A, Geddert H, Faller G . EGFR, KRAS, BRAF-mutations and microsatellite instability are absent in goblet cell carcinoids of the appendix. Pathol Res Pract 2014;210:274–278.
Ramnani DM, Wistuba II, Behrens C et al. K-ras and p53 mutations in the pathogenesis of classical and goblet cell carcinoids of the appendix. Cancer 1999;86:14–21.
Mao C, Yang ZY, Hu XF et al. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol 2012;23:1518–1525.
Yuan ZX, Wang XY, Qin QY et al. The prognostic role of BRAF mutation in metastatic colorectal cancer receiving anti-EGFR monoclonal antibodies: a meta-analysis. PLoS ONE 2013;8:e65995.
De Roock W, Claes B, Bernasconi D et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753–762.
Acknowledgements
We thank Petra Meyer and Thomas Wochnig for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Jesinghaus, M., Konukiewitz, B., Foersch, S. et al. Appendiceal goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid are genetically distinct from primary colorectal-type adenocarcinoma of the appendix. Mod Pathol 31, 829–839 (2018). https://doi.org/10.1038/modpathol.2017.184
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2017.184
This article is cited by
-
Clinicopathological features of appendiceal goblet cell adenocarcinoma in Japan: a multicenter retrospective study
Surgery Today (2023)
-
Appendiceal Goblet Cell Carcinoma: Role of Cytoreductive Surgery (CRS) and Hyperthermic Intraperitoneal Chemotherapy (HIPEC)
Indian Journal of Surgical Oncology (2023)
-
Advanced appendiceal goblet cell carcinoids with intestinal obstruction: two case reports
Clinical Journal of Gastroenterology (2020)
-
Amphicrine carcinoma of the stomach and intestine: a clinicopathologic and pan-cancer transcriptome analysis of a distinct entity
Cancer Cell International (2019)