Abstract
In lung adenocarcinoma, canonical EML4-ALK inversion results in a fusion protein with a constitutively active ALK kinase domain. Evidence of ALK rearrangement occurs in a minority (2–7%) of lung adenocarcinoma, and only ~60% of these patients will respond to targeted ALK inhibition by drugs such as crizotinib and ceritinib. Clinically, targeted anti-ALK therapy is often initiated based on evidence of an ALK genomic rearrangement detected by fluorescence in situ hybridization (FISH) of interphase cells in formalin-fixed, paraffin-embedded tissue sections. At the genomic level, however, ALK rearrangements are heterogeneous, with multiple potential breakpoints in EML4, and alternate fusion partners. Using next-generation sequencing of DNA and RNA together with ALK immunohistochemistry, we comprehensively characterized genomic breakpoints in 33 FISH-positive lung adenocarcinomas. Of these 33 cases, 29 (88%) had detectable DNA level ALK rearrangements involving EML4, KIF5B, or non-canonical partners including ASXL2, ATP6V1B1, PRKAR1A, and SPDYA. A subset of 12 cases had material available for RNA-Seq. Of these, eight of eight (100%) cases with DNA rearrangements showed ALK fusion transcripts from RNA-Seq; three of four cases (75%) without detectable DNA rearrangements were similarly negative by RNA-Seq, and one case was positive by RNA-Seq but negative by DNA next-generation sequencing. By immunohistochemistry, 17 of 19 (89%) tested cases were clearly positive for ALK protein expression; the remaining cases had no detectable DNA level rearrangement or had a non-canonical rearrangement not predicted to form a fusion protein. Survival analysis of patients treated with targeted ALK inhibitors demonstrates a significant difference in mean survival between patients with next-generation sequencing confirmed EML4-ALK rearrangements, and those without (20.6 months vs 5.4 months, P<0.01). Together, these data demonstrate abundant genomic heterogeneity among ALK-rearranged lung adenocarcinoma, which may account for differences in treatment response with targeted ALK inhibitors.
Similar content being viewed by others
Main
Cancer accounted for 8.8 million deaths in 2015, with lung cancer comprising approximately one in five (1.69 million) of those deaths, making it the leading cause of cancer mortality worldwide.1 The anticipated number of new cases of lung and bronchial cancer in the United States for 2017 is 222 500 with 155 870 deaths.2 The 5-year survival rate of lung cancer from 2006 to 2012 in the United States was 18% (15% for men and 21% for women). The treatment and prognosis of lung cancer varies considerably based on the specific pathologic findings and subtype. Broadly, lung cancer is divided into small-cell lung cancer that is aggressive (though sensitive to treatment with radiation), and non-small-cell lung cancer that can show a range of behaviors and treatments dependent on other properties of the tumor, including molecular alterations. Non-small-cell lung cancer itself is divided into squamous carcinoma and adenocarcinoma. In lung adenocarcinoma, the canonical EML4-ALK inversion results in a fusion protein containing a dysregulated, constitutively active ALK kinase domain.3, 4, 5, 6, 7, 8 Although evidence of ALK rearrangement only occurs in a minority (2–7%) of cases,9 ~60% will respond to targeted inhibition of ALK by drugs such as crizotinib, ceritinib, alectinib, and brigatinib.10, 11, 12, 13, 14, 15, 16, 17 ALK rearrangement is associated with specific clinical features, including higher incidence among women, younger patients, patients of Asian descent, and non-smokers. Moreover, there are a handful of histo- and cyto-pathologic features associated with ALK rearrangement (acinar structure, higher histologic grade).18, 19, 20 None of these features (alone or in combination) provide a strong enough association to diagnose the chromosomal rearrangement or select patients for testing, however, and ancillary diagnostic tests remain critical for identifying the therapeutic target.
The canonical ALK rearrangement in lung adenocarcinoma is an inversion of the short (p) arm of chromosome 2, with breakpoints in the ALK and EML4 genes (Figure 1). The ALK breakpoint lies most frequently within intron 19 (reference sequence NM_004304.4), though there are rare exceptions; however, the partner breakpoint within EML4 is quite variable, occurring in a variety of introns, most commonly intron 13, 20, and 6 (reference sequence NM_019063.4).4 Other ALK translocation partners (the most common of which is KIF5B) constitute a small but significant proportion of ALK rearrangements in lung adenocarcinoma, and new rearrangement partners are reported regularly.7, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 The tumorigenic properties of ALK fusion proteins derive from constitutive activity of the tyrosine kinase domain of ALK. The 5′ fusion partner provides promoter and initiation sites, and permits constitutive dimerization of the translated protein.3, 7, 32 The canonical EML4-ALK rearrangement is well established as a driver of cancer, and limited data suggest that different rearrangements may vary in their pathogenic properties, even among the canonical fusions.33, 34, 35 In addition, single-nucleotide variants (SNVs) altering the coding sequence of ALK or other genes (such as EGFR) can drive oncogenesis or impart resistance to targeted inhibition, despite the presence of an ALK rearrangement.36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46
Inversion of chromosome 2p results in constitutively active EML4-ALK fusion protein. Structural rearrangements involving the ALK gene drive a number of cancers. In non-small-cell lung cancer, the most common structural rearrangement involving the ALK locus is an inversion of a portion of chromosome 2p, juxtaposing the 5′ portion of EML4 to the 3′ portion of ALK (a). ALK is not normally transcribed in adult lung, but under the control of the EML4 promoter, the EML4-ALK fusion gene is transcribed. Notably, the reciprocal translocation juxtaposing the 5′ portion of ALK to the 3′ portion of EML4 also occurs, but as it retains the ALK promoter, it is not transcribed (indicated in gray). Wild-type ALK is a receptor tyrosine kinase known to drive developmental pathways, particularly in the nervous system (tyrosine kinase domain indicated by dark-green bars). In the presence of ALK ligand, wild-type ALK proteins homodimerize, driving downstream developmental processes such as cellular proliferation (b). Under transcriptional control of the EML4 promoter, the EML4-ALK fusion protein is expressed in lung tissue, and the dimerization domain of EML4 (dark purple bars) permits unregulated dimerization of the TK domain, constitutively activating downstream pathways.
Clinically, targeted anti-ALK therapy is initiated based on evidence of ALK genomic rearrangement as detected by fluorescence in situ hybridization (FISH) performed on interphase cells in formalin-fixed, paraffin-embedded tissue sections. Because of the promiscuous nature of the translocation, a ‘break-apart’ FISH probe is the preferred identification strategy, forming the basis for the original ‘in vitro device companion diagnostic’ (CDx) assay approved by the US-FDA.47, 48 Although break-apart FISH probes are generally considered sensitive, they do not permit differentiation or identification of ALK partner genes or non-canonical breakpoints, raising the possibility that not all aberrant ALK FISH patterns (among those deemed positive) represent actual fusion events responsive to targeted inhibitors. Indeed, ‘non-productive rearrangements’ involving the ALK locus may disrupt the reading frame of one or both genes, fuse with non-coding intergenic DNA, or result in genes in opposing transcriptional orientation (‘antisense rearrangement’). All of these ‘non-productive’ rearrangements would yield a positive test result by FISH, but produce no targetable protein. More complex rearrangements have also been reported.24 In addition to the genomic variation with breakpoints, formalin-fixed, paraffin-embedded FISH carries potential interpretive limitations including overlapping nuclei or partial sectioning of nuclei, yielding a risk of false-positive or false-negative results.49 Notably, lung adenocarcinoma determined to be ‘ALK positive’ by formalin-fixed, paraffin-embedded FISH has only 60% objective response rate to ALK inhibitors.10, 11, 12, 13, 14, 15 Although orthogonal sequencing methods such as rapid amplification of cDNA ends PCR may be used to identify and characterize non-canonical rearrangements, these techniques can be laborious, and are often limited to the research setting with moderate success.15, 24, 50, 51, 52
Recently, immunohistochemistry for ALK protein expression has been approved as an additional companion diagnostic for ALK-rearranged lung cancer.47, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66 Immunohistochemistry detects expression of the active portion of ALK, and therefore should be positive only when a targetable protein is expressed in the tumor. However, immunohistochemistry results may be subject to pre-analytic factors (eg, specimen fixation times, type of fixative, and ischemic time), and interpretive errors. Moreover, ALK immunohistochemistry is not inherently specific for ALK fusion proteins, and could in principle be confounded by overexpression of ALK driven by other mechanisms, including amplification or alternative transcription initiation (ATI) of ALK, rather than a true fusion protein.40, 67, 68
Targeted DNA next-generation sequencing can offer a more detailed analysis of chromosomal rearrangements, including identification of the specific DNA breakpoints and fusion partners and is now commonly included in many so-called ‘cancer gene panels.’ Targeted identification of DNA level rearrangements generally involves designing capture probes spanning intronic regions where chromosomal breakpoints are known to occur or may use ligation-mediated methods.69, 70 In addition to detailed information regarding the structural rearrangement, next-generation sequencing offers opportunities for broader genomic analysis, including the identification of SNVs in EML4-ALK or in other genes potentially contributing to acquired therapeutic resistance. Clinical grade RNA sequencing (RNA-Seq) has recently become available in the clinical setting and can be performed on routine formalin-fixed, paraffin-embedded material. For fusion detection, a major advantage of RNA-Seq over targeted DNA-based sequencing is that it represents an unbiased approach and can theoretically identify any expressed gene rearrangement without the need for construction of probes covering commonly involved introns. In this study, we sought to characterize the genomic heterogeneity of ALK fusion breakpoints in a set of 33 ALK FISH-positive lung adenocarcinomas using DNA sequencing, RNA-Seq, and immunohistochemistry.
Materials and methods
From a set of 1532 unselected, consecutive cancer cases evaluated for ALK rearrangement by interphase FISH from 2012 to 2016 at Washington University (St Louis, MO, USA), we identified 20 cases that were positive for an ALK rearrangement by FISH and had DNA-based targeted next-generation sequencing performed as part of routine patient care; 12 cases retained enough additional material for additional RNA-Seq. A second set of 13 ALK FISH-positive cases with targeted next-generation sequencing performed as part of routine patient care was obtained from the University of Washington (Seattle, WA, USA). Clinical and demographic data for the patient cohort is given in Table 1.
DNA sequencing was performed using either the Comprehensive Cancer Gene Set Version 2 next-generation sequencing assay (Washington University Genomics and Pathology Services, St Louis, MO, USA) for the 20 cases originating at Washington University or the UW-OncoPlex version 5 assay for the 13 cases originating from the University of Washington. The methods and workflow of both assays have been reported previously.71, 72, 73, 74 Raw data (FASTQs) generated by both assays were analyzed using the Washington University analysis pipeline as described below.
For cases originating from Washington University, formalin-fixed, paraffin-embedded tissue specimens were obtained and reviewed by a board-certified pathologist (JNR and EJD) to confirm the diagnosis and select areas of highest percent tumor cellularity and viability (at least 10%). Tissue was collected from the designated regions either by coring the block with a single-use, disposable 1 mm sterile punch, scraping unstained slides, or by collecting scrolls (complete sections) of tissue from the block. Genomic DNA was extracted manually from the submitted tissue using the QIAamp DNeasy blood and tissue kit (Qiagen, Valencia, CA, USA), fragmented to ~200 base pairs (bp) by ultrasonication, end-repaired, A-tailed, and ligated to sequencing adapters, followed by limited-cycle PCR using the Agilent SureSelect library kit (Agilent Technologies, Santa Clara, CA, USA). All libraries were constructed to contain a single sample-specific 7-bp index. Enrichment of the exons of 151 genes, flanking regions, and selected introns, totaling ~600 kb, was performed using custom solution-phase complementary RNA hybrid-capture followed by additional limited-cycle PCR (SureSelect; Agilent Technologies) to yield at least 5 μmol/L concentration for sequencing (Supplementary Table 1). All libraries were captured individually and pooled at equimolar concentrations prior to sequencing. Up to 1000 ng DNA from each sample was used for sequencing (or the entire amount of available DNA extract, for cases with genomic DNA yields of <1000 ng). Sequencing was performed in multiplex on an Illumina (San Diego, CA, USA) HiSeq 2500 instrument with the manufacturer’s protocol for 101-bp paired-end reads. Base calls were made by Cassava 1.8 (Illumina), and Novoalign 2.08.02 (Novocraft Technologies, Selangor, Malaysia) was used to map the reads to the human reference genome (UCSC build hg19, NCBI build 37.2).
For cases originating at the University of Washington, DNA was extracted from formalin-fixed, paraffin-embedded solid tumor tissue samples using the Gentra Puregene DNA Isolation Kit (catalog #158489; Qiagen). H&E-stained slides were reviewed before DNA extraction for all formalin-fixed, paraffin-embedded samples, and when necessary, macrodissection of tumor-containing regions was performed to enrich tumor cellularity. Tumor cellularity was estimated by review of H&E-stained slides. Sequencing libraries were constructed from 500 ng of DNA using KAPA Hyper Prep kits (Kapa Biosystems, MA, USA) and hybridization was performed using the Oncoplex assay with custom Agilent SureSelect probes (Agilent Technologies; Supplementary Table 2). DNA sequencing was performed on a HiSeq 2500 sequencing system (Illumina) with 2 × 101 bp, paired-end reads, or on a NextSeq 500 (Illumina) with 2 × 150 bp, paired-end reads according to the manufacturer’s instructions. Initial read mapping against the human reference genome (hg19/GRCh37) and alignment processing was performed using BWA version 0.6.1 (http://sourceforge.net/projects/bio-bwa/files).
Cases sequenced at both Washington University and University of Washington were analyzed using the same pipeline. SAMtools22 (version 0.1.18-1), and Genome Analysis ToolKit (GATK; version 1.2) were used to call SNVs and small indel variants. Large indels and chromosomal rearrangements (including ALK rearrangements) were identified using the Breakdancer and ClusterFast software packages.69 Integrative Genomics Viewer26 (IGV version 2.0.16 or later) and the Clinical Genomicist Workspace application version 2.0 (CGW; PierianDX, St Louis, MO, USA) were used for visualization and interpretation. Software parameters and commands are included in Supplementary Table 3. Sequencing included probes targeting ALK introns 16–22 in addition to other cancer-related genes.
For RNA-Seq, RNA was first extracted from formalin-fixed, paraffin-embedded blocks using the Ambion Recoverall kit (ThermoFisher) following a 3 h proteinase K incubation at 55 °C. RNA-Seq libraries were generated using a whole transcriptome approach. cDNA was generated from formalin-fixed, paraffin-embedded RNA by applying random primers to rRNA-depleted material using the Illumina TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold according to the manufacturer’s instructions; inputs of 100–1000 ng were used. Final libraries were quantified using the Qubit assay (ThermoFisher) and library quality was assessed with the Agilent 2100 Bioanalyzer. The libraries were sequenced on an Illumina NextSeq 500 instrument with paired-end 150 bp reads. Fusions were detected from the resulting sequences by first aligning the sequences to the hg19 genome using Tophat2 v2.1.1 with the ‘fusion-search’ flag and the s.d. between mate pairs set to 100 bases, the maximum intron length set to 30 000 bases, the minimum distance to call a fusion was 30 000 bases and the fusion anchor length was required to be at least 10 bases. The default parameter values for the software were used for all other parameters. The resulting predicted fusion genes from the ‘fusions.out’ file were filtered using a custom-filtering script that first assigned the fusion breakpoints to genes based on the UCSC knownGenes annotation file. Any fusion breakpoints that were not supported by at least two reads with different start sites were filtered out as false-positives.
Immunohistochemistry for expression of ALK protein was performed by Clarient Diagnostic Services (ALK D5F3; Aliso Viejo, CA, USA). All immunohistochemistry slides were reviewed by one or more authors on the study, pathologists certified in Anatomic Pathology by the American Board of Pathology (JNR and EJD).
Fluorescence in situ hybridization was performed for ALK rearrangement using the Vysis ALK Break Apart FISH Probe Kit (Abbott Laboratories, Abbott Park, IL, USA) according to the manufacturer’s specifications. FISH results were scored in a CAP/CLIA-accredited laboratory by certified laboratory professionals as part of routine patient care.
Results
The 20 cases from Washington University were sequenced to a mean unique depth of 1,174x (range 172–2776) corresponding to a mean unique depth of 1,110x (range 126–2755) within the targeted introns 16–22 of ALK (reference sequence NM_004304.4). The 13 cases from University of Washington were sequenced to a mean unique coverage depth of 632 (range 326–1331) corresponding to a mean of 674 unique reads in intron 19 of ALK (range 351–1363). The 12 cases analyzed by RNA-Seq generated a mean of 10.9 gigabases (Gb; range 10.0–12.2) per case with a mean of 61 096 unique transcripts detected and a mean expression ratio of 0.30.
Of the 33 total cases in the cohort, 27 cases (82%) demonstrated concordance between formalin-fixed, paraffin-embedded FISH, and detection of ALK rearrangement by DNA next-generation sequencing (Table 2). Figure 2 illustrates a representative concordant case, comparing formalin-fixed, paraffin-embedded FISH, DNA next-generation sequencing, and immunohistochemistry (RNA data not shown). Of the discordant cases, several harbored variants thought to be mutually exclusive of ALK rearrangement in lung adenocarcinoma: case numbers 12 and 18 demonstrated KRAS variants (at codons 13 and 12, respectively), and case number 25 demonstrated an EGFR p.L858R variant.18, 75 In all three cases, the variant allele frequency (VAF) was significant, suggesting the variants do not represent minor subclones within the neoplasm (Table 2). Case number 13 was also positive by FISH and negative for rearrangement by DNA next-generation sequencing, but no mutually exclusive variants were identified; however, ALK immunohistochemistry was also negative in this case. Notably, the FISH results in case numbers 12 and 18 are near the minimum threshold recommended by the manufacturer (Table 2). While we cannot exclude the possibility that ALK rearrangements were not detected in these discordant cases for technical reasons, we note the mean coverage in the ALK intronic region in these cases was not statistically different from the coverage of cases in which a rearrangement was detected (751 vs 938, P=0.48, Student’s t-test). Furthermore, the tumor cellularity was not significantly different in the discordant cases compared to cases in which a rearrangement was detected by DNA next-generation sequencing (57 vs 43%, P=0.08, Student’s t-test).
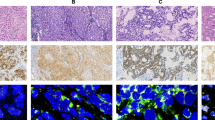
Multiple methodologies identify genomic rearrangements involving the ALK locus in lung adenocarcinoma. A representative case of lung adenocarcinoma (case number 4, H&E in (a) was evaluated with formalin-fixed, paraffin-embedded FISH (b), immunohistochemistry (c), DNA next-generation sequencing (d), and RNA-Seq (data not shown). This patient presented with stage IV adenocarcinoma with metastases to the pleura. Treatment with the first-line anti-ALK agent crizotinib was initiated along with HSP-90 inhibitor as part of a clinical trial. After completion of the study, the patient started single-agent crizotinib. The patient survived 19 months from the time of diagnosis, and was still alive at the time of this study, though with progression based on growth of the pleural metastasis. Formalin-fixed, paraffin-embedded FISH (b) detects genomic rearrangement involving the ALK locus using a break-apart probe strategy. Red and green probes hybridize to genomic positions on each side of the ALK intron 19 breakpoint. When the probes are juxtaposed in the intact chromosome, they yield a yellow signal (filled yellow arrows). Separate red and green signals identify a rearrangement involving the ALK locus (open red arrows). Immunohistochemistry for ALK protein, recently approved by the United States Food and Drug Administration (US-FDA) as an additional methodology, demonstrates robust cytoplasmic staining in neoplastic tissue (c). DNA next-generation sequencing using paired-end sequencing maps multiple sequencing reads from the patient specimen to the reference human genome. (d–f) show a composite screen capture from Integrated Genome Viewer (IGV, available from the Broad Institute), displaying the breakpoint in intron 19 of ALK on the left. Gray bars represent sequencing reads mapping to the reference human genome without mismatch. Sequence reads that map to two different portions of the genome (‘split-reads’ or ‘chimeric reads’) are identified by multicolored bars. These split-reads directly overlie the chromosomal breakpoint. Note that two different patterns of split-reads can be identified, corresponding to the EML4-ALK rearrangement and its reciprocal, ALK-EML4 (see Figure 1 for a schematic explanation of reciprocal rearrangements). Note also that the breakpoints are not identical in the pathologic rearrangement and its reciprocal. Reads that are an unexpected distance from their designated pair (‘unpaired’ or ‘mispaired’ reads; teal and green bars) can be used to locate the rearrangement partner (in this case, EML4 intron 13, shown in e, and the reciprocal partner in intron 3 of the NCKAP5 gene in f). Scale bars represent 200 μm.
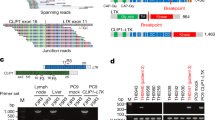
In 12 cases (numbers: 3, 4, 6, 7, 8, 16, 17, 23, 26, 30, 31, and 32; Table 2), both a translocation and a reciprocal rearrangement were identified. In the most straightforward scenario, the reciprocal rearrangement would be the direct inverse of the pathogenic rearrangement (ie, an EML4-ALK rearrangement would be accompanied by the reciprocal ALK-EML4 as in Figure 1a, and case numbers 26, 30, 31, and 32). It is noteworthy, though, that the reciprocal rearrangement does not always involve the exact breakpoints as the primary rearrangement; in some cases, the breakpoint of the reciprocal was markedly distant from the pathogenic breakpoint (see the breakpoints for case numbers 7, 8, 16, 17, and 23 in Table 2). Case number 17 harbors multiple distinct rearrangements involving the ALK locus. Only one of the detected rearrangements appears capable of producing a functional fusion transcript (NBAS-ALK), although an ALK-EML4 rearrangement (rather than the pathogenic EML4-ALK) was also detected (Figure 3). The detection of multiple rearrangements involving the ALK locus has been previously reported in a patient resistant to targeted therapy.44
Multiple independent genomic rearrangements are detected at the ALK locus by DNA next-generation sequencing in a single case of lung adenocarcinoma. Case number 17 is a 55-year-old male non-smoker, stage T1aN2M0 at the time of diagnosis. The patient underwent surgery, then treatment with alectinib for 11 months, continuing at the time of this study. Multiple breakpoints and rearrangement partners were detected within the same specimen, which was collected prior to anti-ALK therapy. (a) depicts one rearrangement involving ALK intron 17. Both the rearrangement and its reciprocal are detected as split-reads; however, only one rearrangement is confirmed by mispaired reads (dark-blue bars). The supported rearrangement has breakpoints in ALK intron 17 (left side) and EML4 intron 21 (right side), a previously unreported rearrangement. It is important to note, though, that the rearrangement supported by this assay represents ALK-EML4, rather than the pathogenic EML4-ALK. (b) depicts a second distinct rearrangement event in intron 19 of ALK (right side), which produces a rearrangement to intron 35 of the NBAS gene. This rearrangement is an interchromosomal translocation between chromosome 2 and its homologous pair, rather than an inversion, and might produce a fusion transcript. The reciprocal rearrangements at this breakpoint involved non-coding intergenic regions on chromosome 2. RNA-Seq did not detect any fusion transcripts from this specimen (data not shown), but immunohistochemistry performed is positive for ALK expression in tumor cells (compare H&E in c to immunohistochemistry in d). Scale bars represent 200 μm.
As noted above, of 33 cases positive for ALK rearrangement by FISH, 27 (82%) showed evidence of rearrangement by DNA sequencing (Table 2). Twenty-one of these rearrangements (91%) correspond with canonical EML4-ALK inversions on chromosome 2 (variant 1—EML4 exon 13 fused to ALK exon 20; variant 2—EML4 exon 20 fused to ALK exon 20; variant 3—EML4 exon 6 fused to ALK exon 20; Figure 4a). In addition, one case demonstrated the less common KIF5B-ALK rearrangement,21, 23 and another case showed a variant isoform of the recently described PRKAR1A-ALK rearrangement26 (Figure 4b).
Five putative ALK fusion transcripts are identified by next-generation sequencing. Among the 33 cases positive for ALK rearrangement by formalin-fixed, paraffin-embedded FISH, we identified seven genomic rearrangements capable of producing fusion transcripts. All putative fusions include the C-terminal region of ALK (green), including the kinase domain (dark-green bars). Three canonical EML4-ALK fusions were identified (a): variant 1 (EML4 exon 13: ALK exon 20; GenBank AB274722.1), variant 2 (EML4 exon 20: ALK exon 20; GenBank AB275889.1), and variant 3 (EML4 exon 6: ALK exon 20; GenBank AB374361.1). All three EML4-ALK fusions include the N-terminal dimerization domain of the EML4 protein (dark-violet bars). Two less common but previously reported fusions were also identified (b): variant 1 of KIF5B-ALK (KIF5B exon 24: ALK exon 20; GenBank AB462413.1), and a previously unreported variant of PRKAR1 A-ALK (PRKAR1A exon 10: ALK exon 20, referred to here as variant 2). Dimerization domains for KIF5B and PRKAR1A are indicated by the dark-orange and dark-blue bars, respectively. Two cases harbored genomic rearrangements predicted to produce novel fusion transcripts (c): SPDYA exon 10: ALK exon 20 and NBAS exon 35: ALK exon 20. Whether or not SPDYA or NBAS would permit constitutive dimerization is unclear. Notably, the NBAS-ALK rearrangement was detected in a complex case with multiple rearrangements. The proportion of detected rearrangements is approximately consistent with the frequency of fusion isoforms in published literature and clinical databases (d; COSMIC). RNA-Seq on a subset of cases confirms fusion partners and quantifies expression of 3′ ALK in transcripts per million reads (TPM; e). Comparison to TCGA data (5 cases with an identified fusion (+), 510 cases with no identified fusion (−)) reveals case number 3 has elevated expression of ALK without evidence of a fusion transcript (case number 3 is highlighted by a purple ring).
Entirely novel rearrangements were identified in four cases (35%). Sequence data from these cases suggest only two of these cases (case numbers 17 and 19) would produce a viable fusion transcript (NBAS-ALK in case 17 and SPDYA-ALK in case 19; Figure 3 and Figure 4c). The remaining rearrangements (case numbers 1 and 6) appear to result in antisense rearrangements, and would not be predicted to produce a fusion transcript (Table 2 and Figure 5). In light of the fact that reciprocal rearrangements are not necessarily identical to the primary pathogenic rearrangement, antisense rearrangements could simply be reciprocals for which the primary rearrangement was present but not detected. In case number 1, only a single rearrangement was detected, but it involves the 3′ portion of ALK as expected from a primary rearrangement rather than a reciprocal. In case number 6, both a primary and reciprocal were detected and neither one would be expected to produce a fusion transcript. Notably, both cases number 1 and number 6 were sampled for sequencing after targeted anti-ALK treatment had been initiated.
DNA next-generation sequencing predicts an antisense rearrangement at the ALK locus. Case number 1 is a 39-year-old non-smoking male who presented with lung adenocarcinoma stage T2aN3M1 (metastasis to the brain). The patient began cisplatin, pemetrexed, and bevacizumab along with whole-brain radiation. The patient was started on crizotinib for 6 months, during which time the radiotherapy was completed. The patient was transitioned to ceritinib for 1 month, then alectinib for 4 months, on which he continues to be stable. Notably, DNA next-generation sequencing was performed on a sample taken during his treatment with crizotinib. Next-generation sequencing showed a single rearrangement involving intron 3 of the ASXL2 gene (a), and intron 20 of ALK (b). Analysis indicated this rearrangement is an inversion of chromosome 2, but as the genes are normally oriented in the same direction, an inversion is predicted to result in an antisense rearrangement, and no transcript (c; compare to Figure 1a). RNA-Seq, in contrast, identified a canonical EML4-ALK variant 1 transcript (data not shown). One possible explanation for the discrepancy is that the tumor harbors multiple rearrangements (as in case number 19), and sampling of a heterogeneous neoplasm yielded different results. The fact that the patient was not naive to treatment lends some support to this hypothesis, as multiple rearrangements in lung adenocarcinoma have been previously described in a patient who developed therapeutic resistance.44
Additional material was available for RNA-Seq of 12 cases. RNA-Seq detects fusion transcripts as well as expression levels of mRNA transcripts (in transcripts per million reads or TPM, Figure 4e), and can offer orthogonal confirmation of FISH or DNA next-generation sequencing. Like DNA next-generation sequencing, RNA-Seq can elucidate some variants conferring acquired therapeutic resistance.44 However, unlike targeted DNA sequencing, RNA-Seq is unbiased, involving no upfront gene enrichment. In 10 of the 12 cases (83%), RNA-Seq data were entirely consistent with the DNA next-generation sequencing data, including identification of the specific isoform of the fusion transcript (Table 2). Case number 3 showed overexpression of ALK mRNA, but no fusion transcript was identified, despite the detection of EML4-ALK genomic rearrangement by DNA next-generation sequencing. The discrepancy may be explained by the quality of the RNA input; for case number 3, the percentage of reads mapping to exons was low (20%; mean 33%, range 20–40%), whereas the percentage of reads mapping to intergenic sequence was high (40%; mean 17%, range 10–40%), suggesting possible contamination by DNA. In case number 1, RNA-Seq detected a canonical EML4-ALK exon13:20 fusion transcript (GenBank: AB274722.1), whereas DNA next-generation sequencing detected a novel, antisense rearrangement predicted to result in no functional transcript (Table 2, Figure 4e, and Figure 5). Notably, case number 1 was sequenced during (rather than prior to) tyrosine kinase inhibitor treatment, and multiple rearrangements have been reported consequent to treatment.44 It is possible, then, that case number 1, like case number 17, harbors multiple independent rearrangements; moreover, the bioinformatic tools implemented to detect chromosomal rearrangement from DNA next-generation sequencing may have different sensitivities from those used in RNA-Seq, potentially overlooking a clone, especially one repressed by therapy.
Immunohistochemistry for expression of ALK protein was performed on 19 cases with available tissue (Table 2). Robust cytoplasmic staining was evident in 17 of the cases (89%). Case number 13 was negative by immunohistochemistry, and case number 12 demonstrated indeterminate positivity (questionable positivity in a handful of cells from a cytology specimen lacking good architecture). Both of these cases had been negative for ALK rearrangement by both DNA next-generation sequencing and RNA-Seq; moreover, case number 12 had a KRAS p.G13C variant (VAF 73%), suggesting the focal immunohistochemistry staining represents false-positivity. Overall, sequencing data suggested three cases should not express ALK protein based on absence of a detectable rearrangement (case numbers 6 and 12) or the orientation of translocation breakpoints (case number 13), but only case number 13 was definitively negative (case number 12 was indeterminate). Case number 6 was positive for ALK expression by immunohistochemistry, despite targeted DNA next-generation sequencing and RNA-Seq showing neither evidence of genomic rearrangement, nor increased transcripts.
Where available, the clinical history of each patient was evaluated with particular attention paid to treatment with targeted ALK inhibitors (Supplementary Table 4). Only 23 of 33 patients (70%) with follow-up clinical data received treatment with a targeted ALK inhibitor (crizotinib, ceritinib, alectinib, or X396) despite positive FISH results. Three patients (case numbers 1, 3, and 9) were sequenced after the initiation of anti-ALK targeted therapy. Two patients (case numbers 8 and 18) were treated for 1 month or less. Among the patients who received targeted therapy and were sequenced prior to the initiation of therapy, average survival was 15.1 months (range: 1–37 months). To assess the clinical utility of the additional information provided by next-generation sequencing, patients who were sequenced prior to beginning anti-ALK therapy were dichotomized based on whether next-generation sequencing detected a canonical EML4-ALK rearrangement or detected another result (no rearrangement or non-canonical/novel rearrangements). Patients harboring next-generation sequencing confirmed, canonical EML4-ALK rearrangements survived an average of 20.6 months (n=14; range=3–42 months). In contrast, patients without canonical EML4-ALK rearrangements survived an average of 5.4 months on anti-ALK therapy (n=6; range=1–11 months), a statistically significant difference (P<0.01 by Student’s t-test). Kaplan–Meier survival curves are shown in Figure 6 demonstrating a significant survival difference between patients with canonical EML4-ALK rearrangements and those with non-canonical rearrangements or without a rearrangement confirmed by next-generation sequencing (P=0.011 and P=0.003, respectively, by Gehan–Breslow–Wilcoxon test (GBWT)).
Next-generation sequencing segregates patients by response to targeted therapy. Of the 33 patients included in the study, 20 patients received targeted therapy directed at ALK, and had next-generation sequencing genomic analysis performed prior to treatment (Supplementary Table 4). The mean survival for these 20 patients is 16.1 months (range 1–42), with the survival curve depicted in a. These 20 patients can be further subdivided based on the results of next-generation sequencing into patients with next-generation sequencing confirmed canonical EML4-ALK rearrangements (n=14; mean survival 20.6 months; range=3–42 months), patients with non-canonical ALK rearrangements (n=3; mean survival 7 months; range=1–11 months), or no rearrangement at all (n=3; mean survival 3.8 months; range=1–8 months). The survival curves of these groups are compared in b, and confirm a statistically significant difference in overall survival between those patients harboring an EML4-ALK rearrangement and those patients who do not (P=0.002 by Gehan–Breslow–Wilcoxon test). The differences in mean survival between patients with confirmed EML4-ALK and those with either non-canonical rearrangements or no rearrangements detected by next-generation sequencing are also each statistically significant (P=0.011 and P=0.003, respectively, by Gehan–Breslow–Wilcoxon test). There is no significant difference in overall survival between patients with non-canonical rearrangements and those with no rearrangement (P=0.67 by Gehan–Breslow–Wilcoxon test), although the number of patients in each group is quite low.
Discussion
EML4-ALK rearrangement, despite detection in only a small proportion of lung adenocarcinoma, is an important predictor of response to targeted therapies. Using DNA sequencing, RNA-Seq, and protein immunohistochemistry to evaluate ALK FISH-rearranged cases, we demonstrate a significant degree of genomic heterogeneity in ALK-rearranged non-small-cell lung cancer. We note that previous studies have found only 60% of patients with evidence of an ALK rearrangement by FISH achieve an objective response to ALK inhibitors10, 11, 12, 13, 14, 15 and that a similar percentage of cases in our study appear to harbor EML4-ALK rearrangements confirmed by next-generation sequencing (21/33 cases or 64%). Moreover, we show that among patients treated with anti-ALK targeted therapy, overall survival (OS) is increased among those with canonical EML4-ALK rearrangements compared to those with novel, or no rearrangements detected by sequencing. It is therefore possible that genomic heterogeneity of ALK fusion breakpoints accounts for some of the difference in treatment response observed in other studies.
There are technical limitations in formalin-fixed, paraffin-embedded FISH (‘technical false-positives’), including cutting artifact, atypical pattern, and interpretive bias.49 There is also biologic variation, potentially yielding therapeutic non-response (‘biologic false-positives’) including ‘non-productive rearrangements,’ multiple rearrangements, resistance variants, and driver variants in other pathways, among others. Although this study focuses on evaluation of ALK-FISH-positive lung adenocarcinoma and therefore cannot address molecular correlation in ALK FISH negative lung adenocarcinoma, a recent study by Ali et al26 compared FISH to a similar DNA sequencing method, and demonstrated a substantial improvement in ALK fusion detection rates. Immunohistochemistry for ALK protein is meant to remedy some of these shortcomings by specifically detecting cases expressing targetable ALK protein, regardless of the underlying genomic changes. Immunohistochemistry comes with its own interpretive challenges and potential confounders, and itself elides relevant underlying biologic variation. Next-generation sequencing-based testing, therefore, offers a number of potential advantages as a diagnostic or confirmatory test for genomic rearrangements.
Targeted DNA next-generation sequencing specifically identifies breakpoints and rearrangement partners, as well as identifying variants in other genes included in the testing panel. From our data, DNA next-generation sequencing provided additional clinically actionable information beyond what formalin-fixed, paraffin-embedded FISH offered in nine cases (27%). In four FISH-positive but DNA next-generation sequencing-negative cases, activating SNVs were detected in other genes (two in KRAS, one in EGFR). Previous studies have shown that these mutations rarely co-occur with ALK gene rearrangements,18, 75 suggesting that the FISH results in those cases might represent false-positives (or alternatively subclones of a heterogeneous neoplasm). In six additional cases, DNA next-generation sequencing identified rare or previously unreported genomic rearrangements involving the ALK locus: KIF5B-ALK, an unreported isoform of PRKAR1A-ALK, a novel SPDYA-ALK rearrangement, a complex rearrangement including a novel NBAS-ALK rearrangement, and two rearrangements predicted to be ‘non-productive,’ resulting in no fusion transcript. On the basis of FISH alone, all of these cases are eligible for targeted anti-ALK therapy, but the DNA next-generation sequencing data suggest these cases may not respond. In all, only 21 of 33 (64%) FISH-positive cases were positive for canonical EML4-ALK rearrangements by high-coverage targeted DNA next-generation sequencing, a proportion similar to the reported objective response rate to targeted ALK inhibition.10, 11, 12, 13, 14, 15 In our data set, DNA next-generation sequencing identified the specific translocation (or provided evidence of mutually exclusive variants) in 95% of cases compared to alternative assays such as rapid amplification of cDNA ends PCR with an estimated sensitivity of 50–70%, depending on the design.15, 24, 50, 51
Targeted DNA next-generation sequencing clearly offers more detailed information about breakpoint structure than alternative methodologies. The additional information necessitates significantly more interpretation. Formalin-fixed, paraffin-embedded FISH and immunohistochemistry ultimately yield a binary result for the detection of rearrangement involving the ALK locus. In contrast, interpretation of DNA next-generation sequencing data may require characterization of breakpoints and identification and characterization of SNVs in ALK as well as in other genes. Furthermore, a novel genomic rearrangement potentially requires understanding the transcriptional orientation of the respective genes as well as the function and properties of the fusion partner, as dimerization has been reported to be a key component of constitutive activation of the EML4-ALK fusion protein.3, 7, 32
Novel and complex chromosomal rearrangements can be particularly challenging to interpret (for example, case number 17 in Table 2 and Figure 4). Although it may appear that the 3′ domain of ALK is fused to inactive DNA, nearby genes may be capable of initiating transcription or splicing, resulting in a viable transcript. Cryptic promoters and splice sites, currently difficult to detect in the routine interpretation of next-generation sequencing data, can also result in a transcript, as well as the under-appreciated phenomena of intergenic splicing76 and alternative transcript initiation.68 RNA-Seq directly identifies fusion transcripts and quantifies transcript expression, offering a more direct test of clinically relevant ALK variation. In our data, RNA-Seq and DNA next-generation sequencing demonstrate a high degree of concordance for detection of rearrangement (92%). In one case, RNA-Seq identified a fusion transcript not detected by DNA next-generation sequencing. However, in general the main drawback of RNA-Seq is the notorious lability of RNA, especially from formalin-fixed, paraffin-embedded tissue, for which pre-analytic specimen processing can significantly alter results.
In principle, ALK immunohistochemistry offers several advantages over other methodologies: it directly detects the highly stable targetable protein, and is a comparatively simple/rapid technique already available to many anatomic pathology laboratories. In practice, though, immunohistochemistry can be inconsistent, resulting in technical errors due to fixation time or inter-observer variability. Our data suggest that, just as with formalin-fixed, paraffin-embedded FISH, immunohistochemistry masks underlying relevant biologic variability. Case number 17 is illustrative—immunohistochemistry is positive, likely due to the novel NBAS-ALK fusion, but does not reflect the multiple breakpoints detected by DNA next-generation sequencing, a feature that has been associated with acquired therapeutic resistance.44 Of the testing methods compared in this study, only DNA next-generation sequencing would accurately capture the potential therapeutic resistance in this case.
The relative advantages and disadvantages of formalin-fixed, paraffin-embedded FISH, DNA next-generation sequencing, RNA-Seq, and immunohistochemistry are summarized in Table 3. Ultimately, though, the salient question is whether a particular testing methodology more accurately predicts response to targeted ALK inhibition. For this reason, knowledge of clinical outcomes is essential. Among patients tested with both FISH and next-generation sequencing prior to receiving anti-ALK therapy (n=20), those patients with the canonical EML4-ALK rearrangement confirmed by next-generation sequencing showed increased mean survival compared to those patients whose next-generation sequencing results showed no rearrangement or a novel/non-canonical rearrangement (20.6 months compared to 5.4 months, respectively, P<0.01). Further analysis of ALK FISH-positive patients without detectable rearrangements by sequencing compared to those with non-canonical rearrangements shows no significant difference between the two groups (P=0.67 by GBWT; Figure 6). Although the number of patients in each category is quite low (three in each category), these data raise the possibility that EML4-ALK rearrangements confirmed by next-generation sequencing respond better to treatment than those not confirmable by next-generation sequencing.
These data are consistent with the idea that canonical ALK fusions detected by sequencing-based methods have better response to anti-ALK therapy; however, our patient cohort is difficult to comprehensively assess for clinical outcomes due to several limitations. First, the number of patients is small, and therefore the study cannot be controlled for either demographic factors (age, ethnicity, and so on) nor for differences in biology (different EML4-ALK isoforms, novel fusion partners, and so on). Moreover, the survival analysis was performed retrospectively and not as part of a well-controlled clinical trial; many of the patients in the study were not started on targeted anti-ALK therapy despite positive ALK FISH results, and many had advanced disease or were not therapeutically naive at the time of initial testing; their tumors may have developed resistance mechanisms or other genomic changes prior to the detection of ALK rearrangement. Therefore, this study cannot definitively address anti-ALK therapeutic response based on sequencing data. Larger, controlled, prospective trials will be needed to more rigorously characterize the exact therapeutic significance of ALK genomic heterogeneity.
Translocation driven cancers are complex and genomic profiling must be approached with a robust understanding of both the underlying biology and the technology used in its assessment. Formalin-fixed, paraffin-embedded FISH and immunohistochemistry are comparatively straightforward to perform and interpret, but belie much of the underlying complexity. Our limited clinical outcome data and the observation that ALK FISH-positive cases harboring mutations thought to be mutually exclusive with ALK rearrangements tested negative by direct sequencing-based methods suggest that ALK FISH has a considerable false-positive rate. In contrast to FISH and immunohistochemistry, next-generation sequencing-based technologies can reveal a great deal more about the tumor—potentially increasing specificity—at the cost of making testing and interpretation more technically challenging and time-consuming. Despite the increased resolution, it is also important not to over-interpret next-generation sequencing results: from our series, 3 cases (numbers 1, 6, and 17) are predicted to generate ‘non-productive’ rearrangements and therefore seem unlikely to respond to tyrosine kinase inhibitor treatment, but at least two of those cases (numbers 1 and 6) do seem to have had a significant response. We note however that both were sequenced after the initiation of anti-ALK therapy, potentially suppressing the founder clone below the limit of detection of next-generation sequencing (Table 2 and Supplementary Table 4). In any case, it is clear that the detection of a ‘non-productive’ rearrangement should be acknowledged, but should not be considered a counter-indication to tyrosine kinase inhibitor therapy. In case numbers 1 and 17, the availability of complementary testing (immunohistochemistry and/or RNA-Seq) offered orthogonal resolution to what might otherwise be challenging data. Optimal testing strategy may best be achieved with different modalities complementing each other, either concurrently or sequentially.
In this study, we demonstrate significant genomic heterogeneity among non-small-cell lung cancer deemed ‘ALK positive’ by FISH. Given such heterogeneity and the advantages conferred by different testing technologies, we suggest multi-modality testing for ALK rearrangement including sequencing-based diagnostics, FISH, and immunohistochemistry. We provide compelling evidence for an increased role for next-generation sequencing-based rearrangement detection in the initial, diagnostic work-up of lung cancer. Advantages of upfront next-generation sequencing-based testing include the detection of chromosome level changes (ALK rearrangements) as well as nucleotide-level mutational data (alternative drivers, resistance variants, and so on). It makes sense to pair assays according to their relative strengths and weaknesses: a more rapid, US-FDA-approved assay (FISH or immunohistochemistry) with a more comprehensive but time-consuming sequencing assay; an assay detecting chromosomal breakage with an assay detecting RNA or protein expression; a directly visualized assay with a sequencing-based assay (ie, if ALK FISH is used for screening, use RNA-Seq for confirmation, and if ALK immunohistochemistry is used for screening, use DNA next-generation sequencing for confirmation; see Supplementary Figures 1a and b).
Although these simple algorithmic approaches are attractive, challenges remain in generalizing clinical laboratory testing. ALK FISH and ALK immunohistochemistry are both US-FDA-approved companion diagnostics, and are likely to persist regardless of other considerations. We also note substantial differences between clinical laboratories in terms how DNA- and RNA-based ALK rearrangement testing is performed. These differences may include sequencing depth, wet-lab target enrichment methods, and bioinformatic analyses that may give rise to differing detection sensitivities and specificities between laboratories. Moreover, as the number of clinically relevant rearrangements in lung cancer increases, targeted DNA-based sequencing rearrangement detection methods (which require targeting of introns) may prove impractical, yielding to methods such as RNA-Seq, Anchored multiplex PCR,77 or whole-genome sequencing.78 Although it may be ideal to perform both RNA and DNA sequencing on every lung adenocarcinoma regardless of ALK FISH or immunohistochemistry status, such an approach may not be economically feasible at present. Two potential testing algorithms are presented in Supplementary Figures 1c and d, attempting to account for differences in the complexity of testing likely to be available in a given laboratory. Given the wealth of information that can be obtained via clinical sequencing-based diagnostics, the precipitous decline in sequencing costs, and the widespread availability of next-generation sequencing-based assays, we expect that such testing will continue to proliferate, and consensus on best practices will emerge.
References
WHO. Cancer Fact Sheet, 2017. Available at: http://www.who.int/mediacentre/factsheets/fs297/en/ (accessed on 23 March 2017).
Society AC. Cancer facts & figures, 2017. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2017.html (accessed on 17 January 2017).
Soda M, Choi YL, Enomoto M et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–566.
Choi YL, Takeuchi K, Soda M et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res 2008;68:4971–4976.
Perner S, Wagner PL, Demichelis F et al. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia 2008;10:298–302.
Martelli MP, Sozzi G, Hernandez L et al. EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am J Pathol 2009;174:661–670.
Hallberg B, Palmer RH . Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 2013;13:685–700.
Rikova K, Guo A, Zeng Q et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190–1203.
Chaft JE, Arcila ME, Paik PK et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther 2012;11:485–491.
Awad MM, Shaw AT . ALK inhibitors in non-small cell lung cancer: crizotinib and beyond. Clin Adv Hematol Oncol 2014;12:429–439.
Friboulet L, Li N, Katayama R et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662–673.
Shaw AT, Kim DW, Nakagawa K et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385–2394.
Antoniu SA. Crizotinib for EML4-ALK positive lung adenocarcinoma: a hope for the advanced disease? Evaluation of Kwak EL, Bang YJ, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-1703. Expert Opin Ther Targets 2011; 15: 351–353..
Shaw AT, Solomon B . Targeting anaplastic lymphoma kinase in lung cancer. Clin Cancer Res 2011;17:2081–2086.
Kwak EL, Bang YJ, Camidge DR et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693–1703.
Peters S, Camidge DR, Shaw AT et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med 2017;377:829–838.
Kim DW, Tiseo M, Ahn MJ et al. Brigatinib in patients with crizotinib-refractory anaplastic lymphoma kinase-positive non-small-cell lung cancer: a randomized, multicenter phase II trial. J Clin Oncol 2017;35:2490–2498.
Inamura K, Takeuchi K, Togashi Y et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol 2009;22:508–515.
Inamura K, Takeuchi K, Togashi Y et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol 2008;3:13–17.
Miyata K, Morita S, Dejima H et al. Cytological markers for predicting ALK-positive pulmonary adenocarcinoma. Diagn Cytopathol 2017;45:963–970.
Ju YS, Lee WC, Shin JY et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res 2012;22:436–445.
Jung Y, Kim P, Jung Y et al. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer 2012;51:590–597.
Takeuchi K, Choi YL, Togashi Y et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 2009;15:3143–3149.
Togashi Y, Soda M, Sakata S et al. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS ONE 2012;7:e31323.
Majewski IJ, Mittempergher L, Davidson NM et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol 2013;230:270–276.
Ali SM, Hensing T, Schrock AB et al. Comprehensive genomic profiling identifies a subset of crizotinib-responsive ALK-rearranged non-small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist 2016;21:762–770.
Hong M, Kim RN, Song JY et al. HIP1-ALK, a novel fusion protein identified in lung adenocarcinoma. J Thorac Oncol 2014;9:419–422.
Fang DD, Zhang B, Gu Q et al. HIP1-ALK, a novel ALK fusion variant that responds to crizotinib. J Thorac Oncol 2014;9:285–294.
Kelly LM, Barila G, Liu P et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci USA 2014;111:4233–4238.
Forbes SA, Beare D, Gunasekaran P et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015;43:D805–D811.
COSMIC, 2017http://cancer.sanger.ac.uk/(accessed on 23 March 2017).
Mano H . Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci 2008;99:2349–2355.
Yoshida T, Oya Y, Tanaka K et al. Differential crizotinib response duration among ALK fusion variants in ALK-positive non-small-cell lung cancer. J Clin Oncol 2016;34:3383–3389.
Lin JJ, Shaw AT . Differential sensitivity to crizotinib: does EML4-ALK fusion variant matter? J Clin Oncol 2016;34:3363–3365.
Armstrong F, Duplantier MM, Trempat P et al. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene 2004;23:6071–6082.
Choi YL, Soda M, Yamashita Y et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734–1739.
Wang YW, Tu PH, Lin KT et al. Identification of oncogenic point mutations and hyperphosphorylation of anaplastic lymphoma kinase in lung cancer. Neoplasia 2011;13:704–715.
Katayama R, Shaw AT, Khan TM et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17.
Sasaki T, Koivunen J, Ogino A et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res 2011;71:6051–6060.
Doebele RC, Pilling AB, Aisner DL et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472–1482.
Hallberg B, Palmer RH . Crizotinib—latest champion in the cancer wars? N Engl J Med 2010;363:1760–1762.
Lin YT, Yu CJ, Yang JC et al. Anaplastic lymphoma kinase (ALK) kinase domain mutation following ALK inhibitor(s) failure in advanced ALK positive non-small-cell lung cancer: analysis and literature review. Clin Lung Cancer 2016;17:e77–e94.
van der Wekken AJ, Saber A, Hiltermann TJ et al. Resistance mechanisms after tyrosine kinase inhibitors afatinib and crizotinib in non-small cell lung cancer, a review of the literature. Crit Rev Oncol Hematol 2016;100:107–116.
Saber A, van der Wekken AJ, Kok K et al. Genomic aberrations in crizotinib resistant lung adenocarcinoma samples identified by transcriptome sequencing. PLoS ONE 2016;11:e0153065.
Ou SH, Klempner SJ, Greenbowe JR et al. Identification of a novel HIP1-ALK fusion variant in non-small-cell lung cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to alectinib. J Thorac Oncol 2014;9:1821–1825.
Kim S, Kim TM, Kim DW et al. Heterogeneity of genetic changes associated with acquired crizotinib resistance in ALK-rearranged lung cancer. J Thorac Oncol 2013;8:415–422.
U.S. Food and Drug Administration. List of approved companion diagnostic devices (in vitro and imaging tools), 2017http://www.fda.gov/medicaldevices/productsandmedicalprocedures/invitrodiagnostics/ucm301431.htm (accessed on 5 March 2017).
Vysis ALK. Break Apart FISH Probe Kit. Abbott Molecular, 2017https://www.molecular.abbott/us/en/products/oncology/vysis-alk-break-apart-fish-probe-kit.
McLeer-Florin A, Lantuejoul S . Why technical aspects rather than biology explain cellular heterogeneity in ALK-positive non-small cell lung cancer. J Thorac Dis 2012;4:240–241.
Takeuchi K, Choi YL, Soda M et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 2008;14:6618–6624.
Weickhardt AJ, Aisner DL, Franklin WA et al. Diagnostic assays for identification of anaplastic lymphoma kinase-positive non-small cell lung cancer. Cancer 2013;119:1467–1477.
Roth A, Streubel A, Grah C et al. A rare case of an EML4-ALK-rearranged lung adenocarcinoma missed by in situ-hybridization but detected by RT-PCR. J Clin Pathol 2014;67:839–840.
McLeer-Florin A, Moro-Sibilot D, Melis A et al. Dual IHC and FISH testing for ALK gene rearrangement in lung adenocarcinomas in a routine practice: a French study. J Thorac Oncol 2012;7:348–354.
Yi ES, Boland JM, Maleszewski JJ et al. Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC score algorithm for FISH. J Thorac Oncol 2011;6:459–465.
Ma H, Yao WX, Huang L et al. Efficacy of D5F3 IHC for detecting ALK gene rearrangement in NSCLC patients: a systematic review and meta-analysis. Oncotarget 2016;7:70128–70142.
Taheri D, Zahavi DJ, Del Carmen Rodriguez M et al. For staining of ALK protein, the novel D5F3 antibody demonstrates superior overall performance in terms of intensity and extent of staining in comparison to the currently used ALK1 antibody. Virchows Arch 2016;469:345–350.
Conde E, Hernandez S, Prieto M et al. Profile of Ventana ALK (D5F3) companion diagnostic assay for non-small-cell lung carcinomas. Expert Rev Mol Diagn 2016;16:707–713.
Savic S, Diebold J, Zimmermann AK et al. Screening for ALK in non-small cell lung carcinomas: 5A4 and D5F3 antibodies perform equally well, but combined use with FISH is recommended. Lung Cancer 2015;89:104–109.
Wynes MW, Sholl LM, Dietel M et al. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK FISH and between evaluators. J Thorac Oncol 2014;9:631–638.
Boland JM, Erdogan S, Vasmatzis G et al. Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Hum Pathol 2009;40:1152–1158.
Paik JH, Choe G, Kim H et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol 2011;6:466–472.
Teixido C, Karachaliou N, Peg V et al. Concordance of IHC, FISH and RT-PCR for EML4-ALK rearrangements. Transl Lung Cancer Res 2014;3:70–74.
Ma H, Yao WX, Huang L et al. Efficacy of D5F3 IHC for detecting ALK gene rearrangement in NSCLC patients: a systematic review and meta-analysis. Oncotarget 2016;7:70128–70142.
von Laffert M, Warth A, Penzel R et al. Multicenter immunohistochemical ALK-testing of non-small-cell lung cancer shows high concordance after harmonization of techniques and interpretation criteria. J Thorac Oncol 2014;9:1685–1692.
Demidova I, Grinevich V, Avdalian A et al. Detection of ALK rearrangements in 4002 Russian patients: the utility of different diagnostic approaches. Lung Cancer 2017;103:17–23.
Pekar-Zlotin M, Hirsch FR, Soussan-Gutman L et al. Fluorescence in situ hybridization, immunohistochemistry, and next-generation sequencing for detection of EML4-ALK rearrangement in lung cancer. Oncologist 2015;20:316–322.
Salido M, Pijuan L, Martinez-Aviles L et al. Increased ALK gene copy number and amplification are frequent in non-small cell lung cancer. J Thorac Oncol 2011;6:21–27.
Wiesner T, Lee W, Obenauf AC et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature 2015;526:453–457.
Abel HJ, Al-Kateb H, Cottrell CE et al. Detection of gene rearrangements in targeted clinical next-generation sequencing. J Mol Diagn 2014;16:405–417.
FusionPlex ALK, RET, ROS1 v2 Kit, ArcherDX, 2017http://archerdx.com/fusionplex-assays/alk-fusion-assay?mid=nav (accessed on 23 March 2017).
Pritchard CC, Salipante SJ, Koehler K et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn 2014;16:56–67.
Pritchard CC, Smith C, Salipante SJ et al. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn 2012;14:357–366.
Cottrell CE, Al-Kateb H, Bredemeyer AJ et al. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn 2014;16:89–105.
Sehn JK, Hagemann IS, Pfeifer JD et al. Diagnostic utility of targeted next-generation sequencing in problematic cases. Am J Surg Pathol 2014;38:534–541.
Zhou JX, Yang H, Deng Q et al. Oncogenic driver mutations in patients with non-small-cell lung cancer at various clinical stages. Ann Oncol 2013;24:1319–1325.
Jividen K, Li H . Chimeric RNAs generated by intergenic splicing in normal and cancer cells. Genes Chromosomes Cancer 2014;53:963–971.
Zheng Z, Liebers M, Zhelyazkova B et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479–1484.
Kohno T, Nakaoku T, Tsuta K et al. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res 2015;4:156–164.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
At the time of this study, RB, PhD, JRA, and JH, PhD were employees of Cofactor Genomics. EJD, MD, is a consultant to Cofactor Genomics. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Rosenbaum, J., Bloom, R., Forys, J. et al. Genomic heterogeneity of ALK fusion breakpoints in non-small-cell lung cancer. Mod Pathol 31, 791–808 (2018). https://doi.org/10.1038/modpathol.2017.181
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2017.181
This article is cited by
-
Deciphering a cell death-associated signature for predicting prognosis and response to immunotherapy in lung squamous cell carcinoma
Respiratory Research (2023)
-
Concomitant novel ALK-SSH2, EML4-ALK and ARID2-ALK, EML4-ALK double-fusion variants and confer sensitivity to crizotinib in two lung adenocarcinoma patients, respectively
Diagnostic Pathology (2022)
-
Reliability analysis of exonic-breakpoint fusions identified by DNA sequencing for predicting the efficacy of targeted therapy in non-small cell lung cancer
BMC Medicine (2022)
-
Kinase Fusion–Related Thyroid Carcinomas: Towards Predictive Models for Advanced Actionable Diagnostics
Endocrine Pathology (2022)
-
Detecting anaplastic lymphoma kinase (ALK) gene rearrangements with next-generation sequencing remains a reliable approach in patients with non-small-cell lung cancer
Virchows Archiv (2022)