Abstract
Over the past decade, comprehensive genomic studies demonstrated that leiomyosarcomas and most of the tumors previously labeled as ‘malignant fibrous histiocytomas’ share complex karyotypes and genomic profiles, and can be referred to as ‘sarcomas with complex genomics’. We recently reported a series of 160 sarcomas with complex genomics such as leiomyosarcomas, myxofibrosarcomas, pleomorphic liposarcomas/rhabdomyosarcomas and undifferentiated pleomorphic sarcomas. These tumors present with a frequent loss of chromosome 10 region encompassing the tumor suppressor gene PTEN. In the present study, we assessed PTEN genomic level and protein expression in this large series of sarcomas with complex genomics, as well as activation of downstream pathways. PTEN partial genomic loss was observed in only 46% of tumors, especially in well-differentiated leiomyosarcomas, whereas up to 68% of these tumors demonstrate a loss of protein expression on western blot analysis. Specific discrepancies in PTEN immunohistochemical results suggested bias in this latter technique. PTEN mutations were rare, with only 4 point mutations in the 65 samples studied. Subsequent activation of AKT and mTOR pathways was only observed in 2 out of 3 of PTEN-deleted tumors. On the other hand, RICTOR, a major component of the mTOR complex 2, was significantly overexpressed in well-differentiated leiomyosarcomas. These results, confirmed on tissue micro-array immunohistochemical analysis of 459 sarcomas, could suggest a link between RICTOR overexpression and leiomyosarcomas oncogenesis. As therapeutics directed against the mTOR pathway are assessed in sarcomas, RICTOR overexpression in sarcomas and its links to therapeutic response need to be assessed.
Similar content being viewed by others
Main
Over the last decade, major changes have occurred in the classification of soft tissue sarcomas. Notably, the subgroup of ‘malignant fibrous histiocytomas’ has been disbanded. Using sharper morphological criteria, most of these poorly differentiated tumors can now be reclassified as poorly differentiated leiomyosarcomas, myxofibrosarcomas, pleomorphic rhabdomyosarcomas and pleomorphic liposarcomas.1, 2 Array comparative genomic hybridization genomic studies sorted out from this group dedifferentiated liposarcomas, characterized by a region of specific amplification encompassing the MDM2 and CDK4 genes.3, 4, 5 On the whole, only 5% sarcomas can still not be classified and are now labeled as ‘undifferentiated pleomorphic sarcomas’.
These genomic studies also demonstrated that, apart from liposarcomas, most of the previously labeled ‘malignant fibrous histiocytomas’ with spindle or pleomorphic morphology share complex karyotypes and genomic profiles.6, 7, 8 They can be thus defined as ‘sarcomas with complex genomics’. Non-supervised clustering analysis of transcriptomic data among these tumors mostly sorted out smooth muscle differentiation genes in well-differentiated leiomyosarcomas without any conspicuous difference between other histological subtypes.9, 10, 11, 12, 13 These results would suggest common complex biological pathways in the oncogenesis of these tumors.
We recently reported a comprehensive genomic study of a large series of 160 sarcomas with complex genomics.13 This study allowed us to evidence frequent chromosome 10q deletions in sarcomas with complex genomics; we could also demonstrate that the tumor suppressor gene PTEN is significantly underexpressed in 10q-deleted tumors. This gene encodes a dual phosphatase protein that can inhibit AKT signaling, implicated in cell proliferation and resistance to apoptosis.14 AKT activates the mTOR pathway, and more specifically, the mTOR complex 1 controlling cell growth and metabolism, whereas the mTOR complex 2 involving the protein RICTOR has roles in cytoskeletal organization and AKT activation.14, 15, 16 Both pathways have been implicated in tumorigenesis.17, 18 In our study, we will focus more specifically on PTEN gene alterations and protein expression in sarcomas with complex genomics, with emphasis on its consequences in activating downstream pathways.
Material and methods
Tumor Samples
This series included the 167 tumor samples (T1 to T167) of our previous series,13 plus 12 additional tumor samples (tumors A01 to A12), for which we had performed an array comparative genomic hybridization analysis, but did not have transcriptomic data. Frozen samples and paraffin blocks were obtained from Pathology Departments throughout France, Switzerland and Canada. Experiments were performed in agreement with the Bioethics Law number 2004-800 and the Ethics Charter from the National Institute of Cancer. Histological typing, tumor grade and smooth muscle differentiation were assessed as previously described.13 Two cell lines were also analyzed, derived from sarcomas with complex genomics samples as previously described.19 LMS148L is derived from T165, a poorly differentiated leiomyosarcoma with myocardin amplification; S19L is derived from an undifferentiated pleomorphic sarcoma (tumor A05).
Array-Comparative Genomic Hybridization and Transcriptomic Analysis
Genomic DNA extraction and array comparative genomic hybridization experiments were performed as previously described.13 Total RNAs from tumor samples were analyzed on GeneCHIP Human Genome U133 Plus 2.0 Array (Affymetrix) and normalized as previously described.13 Hierarchical clustering of tumors T1 to T167 was performed using dChip software (centroid linkage method, rank correlation distance metric, IQR=2).
Real-Time Genomic Quantitative PCR and Sequencing
To determine the copy number status of PTEN and RICTOR, real-time PCR was performed on genomic DNA. Primers and probes for PTEN were designed in intronic sequences to avoid any cross matching with PTEN pseudogene on chromosome 9. The nine exons of the PTEN gene were amplified using intron-based primer pairs. Detailed experimental conditions are supplied in Supplementary Information 1.
Western Blot and Immunohistochemistry
Protocols and antibodies used are described in Supplementary Information 1. On western blot analysis, PTEN protein expression was assessed in a semi-quantitative way (0=no expression, 1=diminished expression, 2=normal expression compared with β-actin signal). Expression of other proteins was assessed by the presence or absence of band at expected size on western blot analysis, except for RICTOR and RAPTOR, where intensity of staining was scored from 0 to 3. On immunohistochemistry, PTEN expression was assessed semi-quantitatively from score 0 (absence of expression) to 1 (decreased expression in tumoral cells compared with normal endothelial cells) and 2 (same intensity of staining in tumoral and normal cells). Slides without signal even in endothelial cells, or with ‘zone effect’ (alternation of entirely positive and entirely negative zones) were considered as ‘non-assessable’. RICTOR immunohistochemical expression was assessed by presence or absence of expression and localization of signal.
Tissue micro arrays were assembled in the Institut Bergonié from 459 mesenchymal tumors issued from various French Pathological Departments. Spots of paraffin-embedded tissue (1 mm) were added in triplicates for each tumor. The 459 tumors were therefore represented by 10 paraffin blocks, each block comprising the same 5 internal controls to ensure reproducibility of results.
Statistical Analysis
Comparisons of non-parametric data were established using Fisher's exact test. Pearsons's correlation factor was used for comparison of quantitative data.
Results
Tumor Samples
The 167 tumor samples previously studied13 were labeled according to their order in the non-supervised clustering analysis, from T1 to T167. As stated previously,13 tumors T21 and T22, T35 and T36, T69 and T70, T85 and T86, T138 and T139 correspond to duplicates of the same tumor, from a different DNA extraction sample. Tumor 118 is the local recurrence of tumor 117, and 144 and 145 are different local recurrences of the same tumor. In this study, well-differentiated leiomyosarcomas clusterized together in group A, and were characterized by an overexpression of smooth muscle-related genes. The other cluster groups were differentiated by overexpression of extracellular matrix, adhesion or inflammation genes (groups C to E), or low expression of genes involved in pericentromeric chromosomal region, mitotic spindle and mitosis control (group B). These groups thus encompass various histological subtypes.
Detailed clinical data, as well as array comparative genomic hybridization, Affymetrix data and mutation analysis regarding PTEN, RICTOR and RAPTOR of the clusterized tumors, the additional samples and the cell lines are displayed in Table 1 . Briefly, the 179 tumor samples analyzed correspond to 172 patients, presenting with 34 well-differentiated leiomyosarcomas, 21 poorly differentiated leiomyosarcomas, 75 undifferentiated pleomorphic sarcomas, 25 myxofibrosarcomas, 9 pleomorphic liposarcomas, and 8 pleomorphic rhabdomyosarcomas. The mean age at diagnosis was 61±16 years old. The sex ratio was 83 females for 87 males, but significantly unbalanced in well-differentiated leiomyosarcomas from cluster A (21 females for 5 males, P=0.0027). Twenty-one tumors (12%) were retroperitoneal, a proportion also significantly higher in well-differentiated leiomyosarcomas from cluster A (P<0.0001). A total of 68% of the tumors (n=110) were of advanced histological grade, in agreement with their poor differentiation.
PTEN Genomic Level
In our series, PTEN array comparative genomic hybridization ratio could be assessed in 111 tumors (Figure 1a). Among them, 60 (54%) displayed a normal PTEN array comparative genomic hybridization ratio (⩾0.8), whereas 43 others (39%) displayed a partial loss (ratio from 0.5 to 0.8) and 8 (7%) displayed a pronounced PTEN loss, with a ratio ⩽0.5, down to 0.28. PTEN partial or complete loss is significantly more frequent in cluster A (P=0.003) and in well-differentiated leiomyosarcomas (P=0.033; Figures 1b and c). PTEN genomic copies could be assessed by genomic quantitative PCR in 53 tumors; the results were correlated with array comparative genomic hybridization data (R=0.780).
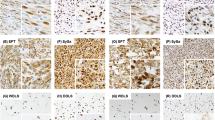
PTEN expression in sarcomas with complex genomics. (a) 10q deletion surrounding PTEN gene in clusterized sarcomas with complex genomics. In this figure, tumors are named and displayed according to their order in non-supervised clustering of Affymetrix data. On the top of the figure, histological type of each tumor is indicated by a color code in filled squares, described in the bottom of the figure. On the graph, array comparative genomic hybridization ratios are symbolized by color dots according to the cyanine 5/cyanine 3 ratios as stated on the bottom of the figure. This figure is a partial reproduction of the Figure 2 of our previous study: new insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics, Gibault L, The Journal of Pathology, 223(1), Copyright 2010 Pathological Society of Great Britain and Ireland; First published by John Wiley & Sons. (b) PTEN array comparative genomic hybridization ratio according to cluster group. (c) PTEN array comparative genomic hybridization ratio according to histological type. (d) PTEN expression on western blot analysis. Each tumor histological type, PTEN array comparative genomic hybridization ratio and PTEN expression on immunohistochemistry is indicated by a color code in filled squares, as defined in the bottom of the figure. (e) Representative cases of immunohistochemistry staining for PTEN, compared with normal endothelial cells (arrows): T06 and T09, normal staining (score 2); A05 and T144 (score 0). T09 and T144 demonstrate discrepancies with western blot and genomic comparative hybridization ratio. Abbreviations: NA, non-assessable, NP, non-performed, RTP, genomic level assessed by real-time genomic PCR, D, discrepancies between immunohistochemistry and western blot/array comparative genomic hybridization analysis; WD-LMS, well-differentiated leiomyosarcomas; PD-LMS, poorly differentiated leiomyosarcomas; UPS, undifferentiated pleomorphic sarcomas; MFS, myxofibrosarcomas; P-LPS, pleomorphic liposarcoma; P-RMS, pleomorphic rhabdomyosarcoma.
PTEN mRNA Expression
Exhaustive Affymetrix data are available on the Gene Expression Omnibus database (GSE23980).
Due to the presence of PTEN pseudogene on chromosome 9, Affymetrix probe sets for PTEN often display a lack of specificity. Two probesets (217492_s_at and 217494_s_at) match both for PTEN and PTENP1. Some of the most specific probesets, 204054_at and 227469_at, displayed low dynamics in our series; furthermore, they display a partial overlap with PTENP1 cDNA when blasted on the University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu/). We therefore analyzed PTEN expression data using 225363_at, a specific probe set matching the non-translated end of PTEN exon 9. As expected, tumors with low array comparative genomic hybridization ratio such as T19, T72 and T93 demonstrate low mRNA expression. For tumors with intermediate or normal array comparative genomic hybridization ratio, there was a good correlation between Affymetrix data and array comparative genomic hybridization ratio (R=0.498), with some notable exceptions, such as cell line S19L that displayed a normal PTEN array ratio without any mRNA expression (detailed figures in Table 1).
Sequencing Analysis
A total of 65 tumor samples were thoroughly analyzed for PTEN mutations. In 10 additional tumors of B cluster group, only a partial sequencing of PTEN gene in exons 6 and 7, and for T40 and T41 in exons 5 and 8, could be assessed, due to a lack of available DNA. Mutations were found in only three tumors. T117 and its local recurrence T118 displayed a punctual deletion in exon 7, with loss of an A base in codon 780, potentially leading to a change in the amino acid sequence from lysine to asparagine on amino acid 260. The frameshift induced would generate a ‘stop’ codon, truncating the protein at amino acid 265. Tumor A05 displayed a punctual mutation at codon 697 (C>T), potentially generating a ‘stop’ codon truncating the protein at amino acid 233. This mutation was also found in the cell line derived from the tumor (S19L).
PTEN Protein Expression
Western blot and immunohistochemical results of the 67 tumors assessed for PTEN protein expression are displayed in Table 2 .
A total of 57 tumors could be assessed for PTEN protein expression by western blot analysis (representative samples in Figure 1d, as well as their correlation with array comparative genomic hybridization and immunohistochemistry results). In this series, 38 tumors displayed a partial (n=28, 49%) or complete (n=11; 19%) loss of PTEN expression, whereas the 18 remaining tumors (32%) retained a normal expression of the protein.
We could retrieve paraffin blocks from 60 tumors for immunohistochemical analysis. In the 52 tumors where PTEN expression was assessable without zone effects, 56% (n=29) retained normal PTEN expression, whereas the others displayed a partial (17%, n=9) or complete (27%, n=14) loss of expression. Western blot and immunohistochemistry results were concordant in 23 out of 43 cases, with discrepancies in 20 cases, western blot results being more correlated with array comparative genomic hybridization status than immunohistochemistry. Representative samples of concordant and discrepant cases are featured in Figure 1e.
Activated Pathways Downstream of PTEN
To evaluate the impact of PTEN loss of expression in sarcomas with complex genomics, we evaluated the activation of AKT and mTOR pathway by assessment of AKT1- and mTOR-specific phosphorylation on western blot analysis in a large series of tumors (Table 2, representative blots in Supplementary Information 2). AKT1 phosphorylation on serine 473 was evidenced in 17 out of 40 tumors, with a strong signal for five tumors; AKT activation thus occurred in 57% (16 out of 28) tumors displaying partial or complete loss of PTEN expression on western blot analysis. Tumors with AKT activation are preferentially clusterized in cluster A, but the difference was not significant. Subsequent mTOR phosphorylation was observed in 11 AKT-activated tumors. LMS148L and S19L only displayed a faint activation of AKT and mTOR pathways.
Components of the mTOR Complexes: RICTOR Expression
In our series, most tumors displayed a gain of chromosome 5p encompassing RICTOR, a gene encoding for the major component of the mTOR complex 2. For each tumor, the mean level of the two array comparative genomic hybridization clones flanking RICTOR was correlated with RICTOR DNA copy number assessed by genomic quantitative PCR (R=0.774; Table 1). Gains superior to 1.2 in RICTOR region were observed in 75 out of 177 tumors (42%; Figure 2a); however, only a few tumors displayed a strong mRNA expression on Affymetrix data, those tumors mostly corresponding to well-differentiated leiomyosarcomas (Figure 2b) from cluster A (Figure 2c). In comparison, RAPTOR gene on chromosome 17 was rarely gained (29 out of 169 tumors, 17%), and its mRNA expression displayed low dynamics.
RICTOR expression in sarcomas with complex genomics. (a) 5p region and RICTOR gene in clusterized sarcomas with complex genomics. In this figure, tumors are named and displayed according to their order in non-supervised clustering of Affymetrix data. On the top of the figure, histological type of each tumor is indicated by a color code in filled squares, described in the bottom of the figure. On the graph, array comparative genomic hybridization ratios are symbolized by color dots according to the cyanine 5 / cyanine 3 ratios as stated on the bottom of the figure. WD-LMS=Well Differentiated leiomyosarcomas, PD-LMS=Poorly Differentiated leiomyosarcomas, UPS=Undifferentiated Pleomorphic Sarcomas, MFS=Myxofibrosarcomas, P-LPS=Pleomorphic Liposarcoma, P-RMS=Pleomorphic Rhabdomyosarcoma. This figure is a partial reproduction of the Figure 2 of our previous study: New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics, Gibault L, The Journal of Pathology, 223(1), Copyright 2010 Pathological Society of Great Britain and Ireland. First published by John Wiley & Sons, Ltd. (b) RICTOR mRNA expression based on the mean level of the Affymetrix probe sets, and its link to the array comparative genomic hybridization ratio. (c) Boxplot analysis of RICTOR Affymetrix expression in each cluster group. (d) RICTOR expression on immunohistochemistry analysis: representative cases. moderate to strong membranous staining in well differentiated leiomyosarcomas such as T16 or T21; ‘dot-like’ paranuclear staining in T20. Absence of staining in most well differentiated leiomyosarcomas and undifferentiated pleomorphic sarcomas (T37, T141), whereas normal smooth muscle cells demonstrate a strong staining in all samples.
Of the 35 tumors that were assessed for RICTOR expression on western blot analysis, only 8 (23%) retained a strong signal, all corresponding to well-differentiated leiomyosarcomas from cluster A or B. This was confirmed on immunohistochemistry, as the group A well-differentiated leiomyosarcomas all displayed a strong membranous expression for RICTOR (except T20 that displayed a ‘dot-like’ paranuclear staining; Figure 2c). Only five tumors that were not well-differentiated leiomyosarcomas displayed RICTOR expression; it was faint and cytoplasmic.
These results were confirmed on a larger series of 459 sarcomas with complex genomics analyzed on tissue micro arrays (exhaustive data in Supplementary Information 3), encompassing 191 leiomyosarcomas, 138 undifferentiated pleomorphic sarcomas, 61 myxofibrosarcomas, 26 pleomorphic sarcomas, 28 dedifferentiated liposarcomas and 15 other sarcomas such as fibrosarcomas or synovial sarcomas. RICTOR was expressed in 91 tumors, its expression was significantly associated with leiomyosarcomas histological subtype (P<0.0001). A total of 77 leiomyosarcomas expressed RICTOR, either in a membranous pattern (n=36), or in a cytoplasmic ‘dot-like’ pattern (n=41). Absence of RICTOR expression in leiomyosarcomas was preferentially observed in grade 3 tumors that are mostly poorly differentiated (P<0.0001). In the other histological subtypes, the rare positive tumors displayed a diffuse cytoplasmic staining, mostly observed in pleomorphic cells. Of note, all normal vascular smooth muscle cells present in samples displayed a strong membranous and cytoplasmic expression for RICTOR and were used as internal positive controls.
Discussion
Partial loss of chromosome 10q is a frequent event in sarcomas with complex genomics.6, 13, 20, 21, 22 Nevertheless, few PTEN mutations have been, to date, described in these tumors. Saito et al23 evidenced 2 mutations in a series of 51 sarcomas with complex genomics, in 2 retroperitoneal leiomyosarcomas. The same team demonstrated an immunohistochemical loss of PTEN in 29% of cases in 48 sarcomas; PTEN protein loss of expression nevertheless did not correlate with the genomic alterations.24 In our series of 65 sequenced cases, we could only evidence two different mutations in four samples (three tumors and one cell line derived from a mutated undifferentiated pleomorphic sarcoma), confirming that PTEN mutation is a rare event in sarcomas with complex genomics. No mRNA expression was observed in these cases, suggesting an effect of the mutation in mRNA stability. Other mechanisms such as PTEN promoter methylation can be discussed in tumors lacking PTEN mRNA expression, despite a normal comparative genomic hybridization ratio; this phenomenon is nevertheless also rare in our series. Several studies have suggested that pten biallelic deletion is not mandatory to induce a pathological phenotype, supporting the concept of haploinsufficiency in pten-deleted murine models.25, 26, 27 This is presumably a frequent event in sarcomas with complex genomics where partial loss of PTEN gene occurs in 39% of tumors, with subsequent partial loss of mRNA and protein expression.
PTEN protein expression has, to date, mainly been assessed by immunohistochemistry, using the 6H2.1 monoclonal antibody, described in carcinomas as the most specific, and correlated to genomic data.28 Yet in our series, discrepancies with western blot and array comparative genomic hybridization results were observed in 20 out of 43 cases. In very rare cases (T115, T130, T144), numerous vessels and endothelial cells retaining PTEN expression in the tumor could potentially explain ‘false positive’ western blot and comparative genomic hybridization results, hindering PTEN actual loss. On the other hand, some tumors like T19, that are homozygously deleted for PTEN and lack mRNA or protein expression on Affymetrix and western blot data, displayed a strong signal on immunohistochemistry, in several independent experiments. This phenomenon was more specifically observed in well-differentiated leiomyosarcomas that all retain a normal immunohistochemical staining, regardless of PTEN genomic status. We also observed this phenomenon in two samples of gastrointestinal stromal tumors not included in this series of sarcomas with complex genomics. This could suggest technical bias such as a non-specific cross reactivity of this antibody in these tumors, maybe with smooth muscle antigens, inciting to a cautious use of this antibody in studies regarding soft tissue sarcomas. Furthermore, immunohistochemistry for PTEN frequently yields to ‘zone effect’ artifacts, hindering immunohistochemical interpretation, as we observed in eight cases in our series. This problem is even more crucial on tissue micro arrays analysis where tumor samples are of very small size.
The study of the PTEN/AKT/mTOR pathway activation we undertook well reflects the genomic complexity of these sarcomas. Although only one-third retain a normal PTEN expression, several tumors with very low levels of PTEN expression did not yield to downstream activation of AKT and mTOR pathways, except in well-differentiated leiomyosarcomas, where AKT subsequent activation is nearly a constant event in PTEN-deleted tumors. Hernando et al29 had already suggested, in a murine model, the implication of the AKT/mTOR complex 1 pathway in the oncogenesis of leiomyosarcomas. This pathway can be blocked by specific inhibitors such as rapamycin.30 Interestingly, we found that RICTOR, the major component of the mTOR complex 2, is specifically overexpressed in well-differentiated leiomyosarcomas, yielding to a strong membranous or cytoplasmic dot-like signal on immunohistochemistry. As it was also observed on immunohistochemistry in normal smooth muscle cells, we can hypothesize that it may have a potential role in smooth muscle differentiation. Indeed, RICTOR has a role in actin polymerization and cytoskeleton organization.31 As actin needs to be polymerized to allow smooth muscle genes transcription,32, 33 RICTOR could have a permissive effect on smooth muscle differentiation of well-differentiated leiomyosarcomas. It may also have a role in leiomyosarcomas oncogenic processes, as it is the kinase responsible for AKT1 phosphorylation on serine 473, thus allowing its full activity.16 RICTOR genomic gains have indeed been linked to tumor recurrence in hepatocellular carcinoma,34 and its overexpression in glioma cell lines lead to increased anchorage-independent growth and motility.35
Thus this study allowed us to demonstrate, in a large series of sarcomas with complex genomics, the frequent loss of PTEN expression in these tumors, presumably by a ‘gene-dose effect’ mechanism. Subsequent AKT and mTOR activation is nevertheless inconstant. Furthermore, immunohistochemistry for PTEN demonstrated technical bias, especially in leiomyosarcomas, precluding its use in pre-therapeutic assessment in these tumors. Indeed, in a small series of temsirolimus-treated leiomyosarcomas, we could not predict therapeutical response by PTEN immunohistochemical assessment.36 Interestingly, our results also point out that the mTOR complex 2 component RICTOR, yet scarcely studied, could have a major role in leiomyosarcomas oncogenesis. As therapeutics using mTOR inhibitors are being assessed in soft tissue sarcomas,36, 37, 38, 39 further comprehensive studies are needed to determine which subgroups of patients would benefit more from such therapeutics. Additional simple predictive tools are needed to assess activation of mTOR pathway in soft tissue sarcomas, as well as RICTOR overexpression and their potential influence in the therapeutic response.
References
Fletcher CD . The evolving classification of soft tissue tumors: an update based on the new WHO classification. Histopathology 2006;48:3–12.
Dei Tos AP . Classification of pleomorphic sarcomas: where are we now? Histopathology 2006;48:51–62.
Chibon F, Mariani O, Derré J, et al. A subgroup of malignant fibrous histiocytomas is associated with genetic changes similar to those of well-differentiated liposarcomas. Cancer Genet Cytogenet 2002;139:24–29.
Chibon F, Mariani O, Mairal A, et al. The use of clustering software for the classification of comparative genomic hybridization data. An analysis of 109 malignant fibrous histiocytomas. Cancer Genet Cytogenet 2003;141:75–78.
Coindre JM, Mariani O, Chibon F, et al. Most malignant fibrous histiocytomas developed in the retroperitoneum are dedifferentiated liposarcomas: a review of 25 cases initially diagnosed as malignant fibrous histiocytoma. Mod Pathol 2003;16:256–262.
Derré J, Lagacé R, Nicolas A, et al. Leiomyosarcomas and most malignant fibrous histiocytomas share very similar comparative genomic hybridization imbalances: an analysis of a series of 27 leiomyosarcomas. Lab Invest 2001;81:211–215.
Larramendy ML, Gentilec M, Soloneskib S, et al. Does comparative genomic hybridization reveal distinct differences in DNA copy number sequence patterns between leiomyosarcoma and malignant fibrous histiocytoma? Cancer Genet Cytogenet 2008;187:1–11.
Carneiro A, Francis P, Bendahl PA, et al. Indistinguishable genomic profiles and shared prognostic markers in undifferentiated pleomorphic sarcoma and leiomyosarcoma: different sides of a single coin? Lab Invest 2009;89:668–675.
Baird K, Davis S, Antonescu CR, et al. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res 2005;65:9226–9235.
Francis P, Namløs HM, Müller C, et al. Diagnostic and prognostic gene expression signatures in 177 soft tissue sarcomas: hypoxia-induced transcription profile signifies metastatic potential. BMC Genomics 2007;8:73.
Nakayama R, Nemoto T, Takahashi H, et al. Gene expression analysis of soft tissue sarcomas: characterization and reclassification of malignant fibrous histiocytoma. Mod Pathol 2007;20:749–759.
Beck AH, Lee CH, Witten DM, et al. Discovery of molecular subtypes in leiomyosarcoma through integrative molecular profiling. Oncogene 2010;29:845–854.
Gibault L, Pérot G, Chibon F, et al. New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J Pathol 2011;223:64–71.
Bellacosa A, Kumar CC, Di Cristofano A, et al. Activation of AKT Kinases in Cancer: Implications for Therapeutic Targeting. Adv Cancer Res 2005;94:29–86.
Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor independent pathway that regulates the cytoskeleton. Curr Biol 2004;14:1296–1302.
Sarbassov DD, Ali SM, Sabatini DM . Phosphorylation and regulation of Akt/PKB by the rictor–mTOR complex. Science 2005;307:1098–1101.
Jiang BH, Liu LZ . PI3K/PTEN signaling in angiogenesis and tumorigenesis. Adv Cancer Res 2009;102:19–65.
Guertin DA, Stevens DM, Saitoh M, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell 2009;15:148–159.
Pérot G, Derré J, Coindre JM, et al. Strong smooth muscle differentiation is dependent on myocardin gene amplification in most human retroperitoneal leiomyosarcomas. Cancer Res 2009;69:2269–2278.
Mandahl N, Fletcher CD, Dal Cin P, et al. Comparative cytogenetic study of spindle cell and pleomorphic leiomyosarcomas of soft tissues: A report from the CHAMP study group. Cancer Genet Cytogenet 2000;116:66–73.
Hu J, Rao UN, Jasani S, et al. Loss of DNA copy number of 10q is associated with aggressive behavior of leiomyosarcomas: a comparative genomic hybridization study. Cancer Genet Cytogenet 2005;161:20–27.
Meza-Zepeda LA, Kresse SH, Barragan-Polania AH, et al. Array comparative genomic hybridization reveals distinct DNA copy number differences between gastrointestinal stromal tumors and leiomyosarcomas. Cancer Res 2006;66:8984–8993.
Saito T, Oda Y, Kawaguchi K, et al. PTEN/MMAC1 gene mutation is a rare event in soft tissue sarcomas without specific balanced translocations. Int J Cancer 2003;104:175–178.
Kawaguchi K, Oda Y, Saito T, et al. Genetic and epigenetic alterations of the PTEN gene in soft tissue sarcomas. Hum Pathol 2005;36:357–363.
Di Cristofano A, Pesce B, Cordon-Cardo C, et al. Pten is essential for embryonic development and tumor suppression. Nat Genet 1998;19:348–355.
Di Cristofano A, De Acetis M, Koff A, et al. Pten and p27/KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet 2001;27:222–224.
Trotman LC, Niki M, Dotan ZA, et al. Pten dose dictates cancer progression in the prostate. PLoS Biology 2003;1:385–396.
Pallares J, Bussaglia E, Martínez-Guitarte JL, et al. Immunohistochemical analysis of PTEN in endometrial carcinoma: a tissue microarray study with a comparison of four commercial antibodies in correlation with molecular abnormalities. Mod Pathol 2005;18:719–727.
Hernando E, Charytonowicz E, Dudas ME, et al. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med 2007;13:748–753.
Chiang GG, Abraham RT . Targeting the mTOR signaling network in cancer. Trends Mol Med 2007;13:433–442.
Sarbassov DD, Ali SM, Sengupta S, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 2006;22:159–168.
Mack CP, Somlyo AV, Hautmann M, et al. Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J Biol Chem 2001;276:341–347.
Posern G, Treisman R . Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 2006;16:588–596.
Villanueva A, Chiang DY, Newell P, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology 2008;135:1972–1983.
Masri J, Bernath A, Martin J, et al. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res 2007;67:11712–11720.
Italiano A, Kind M, Stoeckle E, et al. Temsirolimus in advanced leiomyosarcomas: patterns of response and correlation with the activation of the mammalian target of rapamycin pathway. Anticancer Drugs 2011;22:463–467.
Chawla SP, Tolcher AW, Staddon AP, et al. Survival results with AP23573, a novel mTOR inhibitor, in patients (pts) with advanced soft tissue or bone sarcomas: update of phase II trial [abstract]. J Clin Oncol 2006;24:521.
Okuno S, Mahoney M, Bailey H, et al. A multicenter phase 2 consortium (P2C) study of the mTOR inhibitor CCI-779 in advanced soft tissue sarcomas (STS) (abstract). J Clin Oncol 2006;24:521.
Dancey JE, Monzon J . Ridaforolimus: a promising drug in the treatment of soft-tissue sarcoma and other malignancies. Future Oncol 2011;7:827–839.
Acknowledgements
We warmly thank the French Sarcoma Group pathologists who provided us with frozen tumor samples and paraffin blocks; Dr Nicolas Sévenet who communicated us PTEN primers and sequencing conditions; Dr Frédérique Larousserie for her advice on immunohistochemistry. This work was supported by grants from INSERM, Institut Curie, INCa (PL007), PHRC 2007/05, Conticanet network and Fondation de France. LG was supported by grants from Paris Descartes University and the Assistance Publique-Hôpitaux de Paris. The construction of the human Bacterial Artificial Chromosome-array was supported by grants from the Carte d’Identité des Tumeurs (CIT) program of the Ligue Nationale Contre le Cancer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Exhaustive Affymetrix data are available on Gene Expression Omnibus genomic database (GSE23980). Array-comparative genomic hybridization data are available as excel files upon request.
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Gibault, L., Ferreira, C., Pérot, G. et al. From PTEN loss of expression to RICTOR role in smooth muscle differentiation: complex involvement of the mTOR pathway in leiomyosarcomas and pleomorphic sarcomas. Mod Pathol 25, 197–211 (2012). https://doi.org/10.1038/modpathol.2011.163
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2011.163
Keywords
This article is cited by
-
Using biology to guide the treatment of sarcomas and aggressive connective-tissue tumours
Nature Reviews Clinical Oncology (2018)
-
mTORC2/AKT/HSF1/HuR constitute a feed-forward loop regulating Rictor expression and tumor growth in glioblastoma
Oncogene (2018)
-
Scattered genomic amplification in dedifferentiated liposarcoma
Molecular Cytogenetics (2017)
-
PI3K/AKT/mTOR inhibition in combination with doxorubicin is an effective therapy for leiomyosarcoma
Journal of Translational Medicine (2016)
-
Nouvelles perspectives et biomarqueurs dans le diagnostic et la prise en charge des tumeurs gynécologiques
Oncologie (2012)