Abstract
The development of easily accessible tools for human immunophenotyping to classify patients into discrete disease endotypes is advancing personalized therapy. However, no systematic approach has been developed for the study of inflammatory lung diseases with often complex and highly heterogeneous disease etiologies. We have devised an internally standardized flow cytometry approach that can identify parallel inflammatory alveolar macrophage phenotypes in both the mouse and human lungs. In mice, lung innate immune cell alterations during endotoxin challenge, influenza virus infection, and in two genetic models of chronic obstructive lung disease could be segregated based on the presence or absence of CD11b alveolar macrophage upregulation and lung eosinophilia. Additionally, heightened alveolar macrophage CD11b expression was a novel feature of acute lung exacerbations in the SHIP-1−/− model of chronic obstructive lung disease, and anti-CD11b antibody administration selectively blocked inflammatory CD11bpos but not homeostatic CD11bneg alveolar macrophages in vivo. The identification of analogous profiles in respiratory disease patients highlights this approach as a translational avenue for lung disease endotyping and suggests that heterogeneous innate immune cell phenotypes are an underappreciated component of the human lung disease microenvironment.
Similar content being viewed by others
Introduction
Immunophenotyping, as the process of identifying discrete patient immune cell phenotypes, has gained prominent traction in areas such as HIV and leukemia research where perturbations in immune cell homeostasis directly reflect disease pathogenesis and correlate with patient prognosis.1, 2, 3 Using flow cytometry, immune cell surface markers with bimodal distributions (which are either negative or positive in expression) allow unambiguous discrimination of immune cell profiles that correlate with clinical outcome, such as the presence CD8+INF-γ+ double-positive cells in HIV patients and CD38+ peripheral blood cells in B-cell chronic lymphocytic leukemia patients.1, 2
A potentially valuable yet underutilized area for lung disease immunophenotyping lies in the study of human alveolar macrophages (AMΦs), which are the predominant immune cells within the alveolar airspaces. However, apart from the utilization of CD14 and CD16 to discern between classical CD14+CD16− and patrolling CD14lowCD16+ monocytes (represented by CX3CR1lowGr-1+ and CX3CR1highGr-1− counterparts in mice),4 few macrophage (MΦ) surface markers exist in a convenient bimodal distribution pattern for disease-based characterization. Few are also identically expressed between mice and men.5 This suggests two possibilities: that more translatable MΦ markers remain to be identified or there is an abundance of surface markers with continuous distributions that have rendered themselves inappropriate for traditional immunophenotyping. Considering the depth of study in the field of MΦ heterogeneity, the latter appears as the most likely option.
Over the past few years, personalized therapy of human lung diseases has been greatly advanced by the concept of disease endotypes, which links clinically observed phenotypes to underlying molecular mechanisms.6 Endotyping asthma has successfully identified discrete patient subgroups who share a similar disease progression or treatment response,7 reorganizing patient management through the implementation of more personalized treatment strategies. This asks whether a similar approach can be applied to chronic obstructive pulmonary disease (COPD), which is mechanistically heterogeneous and involves high patient symptom burdens, which manifest independently of severe lung function decline.8, 9 In particular, recent reports of a hyper-inflammatory COPD patient subgroup with increased disease mortality and acute lung exacerbations risk suggest that inflammatory and non-inflammatory disease endotypes may exist in COPD.9, 10 This concept is emphasized by the existence of both immune and non-immune driven mechanisms of emphysema induction in mice.11, 12, 13 Interestingly, in mice, deletion of Src homology 2 domain-containing 5’ inositol phosphatase-1 (Inpp5d or SHIP-1; a negative regulator of growth factor receptor–mediated myeloid cell activation and survival) or constitutive activation of Hematopoietic cell kinase (Hck; a Src-family kinase abundant in myelomonocytic cells) both lead to the spontaneous development of chronic inflammatory lung disease sharing features of human COPD. SHIP-1−/− mice develop AMΦ-driven chronic lung inflammation, severe emphysema,14, 15, 16, 17, 18 and COPD co-morbidities, such as wasting, osteoporosis and an increased susceptibility to bacterial infections.19, 20, 21 HckF/F gain-of-function mice, generated by a tyrosine to phenylalanine substitution (Y499F) in the regulatory domain which prevents Hck inactivation, develop airway mucus metaplasia and mild emphysema characterized by chronic eosinophilic and monocytic lung inflammation and are hypersensitive to lipopolysaccharide (LPS).22 Further study of SHIP-1−/− and HckF/F mouse models may thus help refine the search for hyper-inflammatory COPD endotypes.
To facilitate the discovery of parallel lung immunophenotypes between mice and humans, we devised a new and internally standardized approach using flow cytometry that focuses on AMΦs and other innate immune cell subpopulations as defects in these cell types are linked to asthma, COPD, and recurrent bacterial and viral chest infections. This easily adoptable approach compares CD11b expression levels on AMΦs with anchor CD11b expression levels in blood neutrophils and additionally identifies CD11b AMΦ upregulation as a hallmark of acute lung exacerbations in SHIP-1−/− mice. Overall, our study incorporates AMΦ phenotyping with other preexisting parameters for clinical endotyping (the presence of eosinophilia or neutrophilia) to demonstrate that innate lung immunophenotypes exist heterogeneously in both mouse models of chronic inflammatory lung disease and human patients.
Results
Identification of distinct inflammatory immune cell profiles in mouse lungs
We and others have previously identified residential AMΦ CD11b upregulation as a marker of acute and chronic lung inflammation in mice.15, 23 Residential AMΦs are CD11bneg in healthy adult mouse lungs at homeostasis. Under selective inflammatory conditions, CD11b expression is upregulated and differential CD11bpos AMΦ proportions can distinguish between the peak and resolution of acute lung inflammation, respectively.15 This scenario (either an absence or presence of inflammation-associated CD11bpos AMΦs) is ideal for immunophenotyping as binary CD11b expression negates interpretation ambiguities and excessive standardization procedures. In translating this to human patient samples, however, we discovered that CD11b was basally expressed on all human AMΦs but varied noticeably between individual patients (Figure 1a). As MΦ markers rarely directly translate between man and mouse, we screened for other putative MΦ activation markers such as CD14, CD86, major histocompatibility complex (MHC) Class II, CD124, CD206, Toll-like receptor (TLR)-2, and TLR-4 but observed consistently high or low surface expression levels (data not shown).
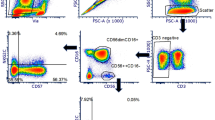
Identification of distinct inflammatory profiles in mouse bronchoalveolar lavage (BAL). (a) CD11b vs. CD11c expression profiles of human alveolar macrophages (AMΦs) compared with healthy adult C57BL/6 mice. Human BAL samples collected and analyzed on separate days, with laser voltages standardized by the use of single peak allophycocyanin (APC) and APC-Cy7 fluorescent beads (blue: stained AMΦs, gray: non-binding antibody (Ab) control). (b) Flow cytometry used to characterize blood neutrophil CD11b expression levels in phosphate-buffered saline vehicle, Day 3 lipopolysaccharide (LPS)-treated, and Day 10 Mem71 influenza virus-infected C57BL/6 mice (n=3, dotted gray: non-binding Ab controls, left panel). Overlay of BAL cell (blue) and blood neutrophil (red) staining with anti-CD11b and anti-CD11c Abs shows distinct inflammatory BAL profiles in individual mice (n=3–6, right panel). AMΦ CD11b upregulation indicated as a rightwards shift of AMΦ CD11b expression with respect to blood neutrophils. (c) Flow cytometry confirms similar CD11b expression on blood neutrophils from SHIP-1−/−, HckF/F, and C57BL/6 mice (n=3, dotted gray: non-binding Ab controls, left panel). Overlay of CD11b vs. CD11c staining of BAL cells (blue) and blood neutrophils (red) in C57BL/6, SHIP-1−/−, and HckF/F mice. (n=6, right panels). (d, e) Hematoxylin and eosin–stained sections of post-BAL inflation fixed lungs from flow cytometry–profiled mice (n=4, bar=100 μm). MFI, mean fluorescence intensity; PMN, polymorphonuclear neutrophil.
This led us to postulate that human AMΦ surface markers (including CD11b) are generally continuous rather than bimodal in distribution, especially as human AMΦs are under constant exposure to a diverse repertoire of inhaled insults and environmental antigens. Unfortunately, flow cytometry measurement of markers with a continuous distribution loses quantitative meaning outside single experiments as expression is presented as a mean fluorescence intensity (MFI), a value which changes with cell autofluorescence, antibody (Ab) signal strength, and laser voltage settings. As MFI values are relative, large-scale clinical usage becomes limited even with single peak autofluorescent bead set-ups, as bead values still fluctuate within a small range between experiments and may diminish over time, hampering diagnostic accuracy. Thus, with continuous AMΦ CD11b expression as our marker of interest, we explored whether alternative analytical methods could still detect distinct lung immune cell profiles in both mice and humans using flow cytometry. To do this, we re-examined established mouse models of acute and chronic lung inflammation for potential internal anchors of AMΦ CD11b expression (gating strategy in Supplementary Figure S1 online).
We observed that blood neutrophil CD11b expression was surprisingly constant between phosphate-buffered saline (PBS) vehicle and LPS-challenged or Mem71 influenza virus–infected mice independent of lung pathology (Figure 1b and Supplementary Table S1 online). Similarly, we found that blood neutrophil CD11b expression remained constant in mice that spontaneously developed inflammatory lung disease (SHIP-1−/− and HckF/F mice; Figure 1c and Supplementary Table S1 online). In acute lung challenge studies, day 3 post-LPS challenge and day 10 post-influenza virus infection were chosen as they represent the peak of acute lung inflammation and initiation of disease resolution after virus is cleared, respectively, in these two different inflammatory conditions. In contrast to Mem71 infection, resolution at day 10 post-LPS challenge is characterized by a restoration to the homeostatic CD11bneg AMΦ phenotype.15 In mice, blood neutrophils were CD11bpos with a staining intensity between the expression range of CD11bneg and CD11bhighCD11chigh AMΦs, providing an internal reference point to which AMΦ CD11b expression could be compared (Figure 1b,c). This approach negates influences from Ab signal strength and laser voltage settings on MFI level, as the ratio of AMΦ to blood neutrophil CD11b expression remains internally constant (see Supplementary Figure S2a and Supplementary Table S1 online). Differences in AMΦ CD11b expression and the appearance of CD11bhighCD11cpos or CD11bposCD11cneg cell subsets were observed in bronchoalveolar lavage (BAL) from C57BL/6 mice following LPS challenge and Mem71 influenza virus clearance and in SHIP-1−/− and HckF/F mice (see Supplementary Figure S2b,c and Supplementary Table S1 online).16, 17, 22 Increased AMΦ to blood neutrophil CD11b expression profiles were also observed for increasing LPS doses at day 3 post-challenge in C57BL/6 mice (see Supplementary Figure S2d online). Critically, distinct BAL cell profiles could be identified in animal models with different lung tissue histopathology (Figure 1d,e).
Further characterization of mouse immune cell subpopulations
Further characterization of CD11bhighCD11cpos immune cells (subset M) in mouse BAL profiles (Figure 2a) showed that these were lung monocytes with increased MHC Class II, CD14, and CD86 surface expression compared with AMΦs (Figure 2b). MHC Class II is heterogeneously expressed on lung monocytes but not residential AMΦs in different inflammatory lung conditions, emphasizing the selectivity of CD11b as a heterogeneously expressed marker of residential AMΦs (see Supplementary Figure S2e online). A second subset of CD11cnegCD11bpos immune cells (subset E; Figure 2a) was also present in Mem71-infected and HckF/F mouse BAL, which stained negatively for myeloid markers Class II, CD14, or CD86 (Figure 2b). Cell differentials using BAL cytospins, cell sorting, and Siglec F staining confirmed these cells as Siglec Fpos lung eosinophils as previously reported (see Supplementary Figure 3S online and Figure 2c; granular cells with eosinophilic cytoplasm as denoted by white arrows).24
Further characterization of immune cell subgroups in heterogeneous mouse bronchoalveolar lavage (BAL) profiles. (a) CD11b vs. CD11c staining of BAL cells segregates CD11chighCD11blow-high residential alveolar macrophages (AMΦs; subset R or Ri and Rii in SHIP-1−/− mice), CD11cposCD11bhigh lung monocytes (subset M), and CD11cnegCD11bpos lung eosinophils (subset E; sorted and characterized in Supplementary Figure S3 online). Representative of n=3–6 (b) Class II, CD14, and CD86 expression of gated immune cell subsets (dotted gray: non-binding antibody control). Representative of n=3–6. (c) BAL cytospins stained using Hemacolor (Merck Millipore). Eosinophils appear as polymorphonuclear leukocytes with pink-red cytoplasmic staining (white arrows; representative of n=4, bar=20 μm).
Identification of new immune cell parameters associated with acute lung exacerbations
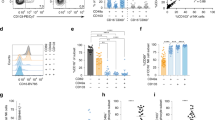
Acute exacerbations of COPD classically involve increased neutrophil recruitment and pro-inflammatory cytokine production,25, 26 although eosinophils have also been increasingly linked to exacerbation predisposition.27 However, it remains unknown how conserved or heterogeneous this inflammatory milieu is between affected patients. SHIP-1−/− and HckF/F mice have spontaneous inflammatory and emphysematous phenotypes that resemble patterns observed in human COPD.16, 17, 22 To test whether specific BAL profiles could also be discerned during COPD-like acute lung exacerbations, SHIP-1−/− and HckF/F mice were challenged with a single intranasal dose of LPS. LPS-challenged SHIP-1−/− and HckF/F mice both developed exacerbated lung pathology resembling acute lung pneumonia by day 3 post-LPS challenge (Figure 3). In SHIP-1−/− mice, acute exacerbations were characterized by CD11bhighCD11cpos monocyte recruitment and further AMΦ CD11b upregulation (Figure 3a–c,m). Exacerbation severity as evidenced by increased lung hemorrhaging, posture changes, piloerection, and increased respiratory effort was variable (data not shown) and correlated with the degree of AMΦ CD11b upregulation visualized as a rightwards signal shift of AMΦ CD11b expression compared with blood neutrophils (Figure 3c). Neutrophil, monocyte, and T lymphocyte numbers were also increased in LPS-challenged SHIP-1−/− mice (Figure 3d–f). AMΦ CD11b upregulation and lung monocyte recruitment were also characteristic of LPS-challenged HckF/F mice (Figure 3i,n). Upregulated AMΦ CD11b expression was modest compared with challenged SHIP-1−/− mice and did not reach statistical significance (Figure 3i,n, *P=0.087). Heightened lung neutrophil recruitment was the dominant feature of challenged HckF/F mice, with no alterations in lung eosinophil numbers (Figure 3j). Interestingly, lung tissue and immune cell apoptosis was found in HckF/F but not SHIP-1−/− acute lung exacerbations (see Supplementary Figure S4 online). Altogether, these results demonstrate that increased CD11b AMΦ expression is a mouse marker of active lung inflammation with differential patterns of AMΦ CD11b upregulation and eosinophil, neutrophil, or monocyte recruitment present in acute and chronic inflammatory lung conditions as well as COPD acute exacerbation models.
Distinct inflammatory signatures appear in mouse chronic obstructive pulmonary disease models of acute lung exacerbations. (a) Inflation fixed lungs from C57BL/6 and SHIP-1−/− mice day 3 post-lipopolysaccharide (post-LPS) or phosphate-buffered saline vehicle challenge (bar=1 cm). (b) Hematoxylin and eosin (H&E)-stained sections of inflation fixed lungs (n=4, bar=100 μm). (c) Overlay of CD11b vs. CD11c staining from bronchoalveolar lavage (BAL) cells and blood neutrophils of day 3 LPS-challenged SHIP-1−/− mice (blue: BAL cells, red: blood neutrophils; n=3–7). Mice were scored as moderate or severe responders based on pneumonia severity, postural changes, piloerection and increased respiratory effort. (d–f) Flow cytometry used to calculate total alveolar macrophage (AMΦ), monocyte, neutrophil (polymorphonuclear neutrophils (PMNs)), and lymphocyte numbers (n=4–7, *P<0.05, **P<0.01, two-tailed Student’s t-test). (g) Inflation fixed lungs from HckF/F mice at day 3 post-LPS challenge (bar=1 cm). (h) H&E sections of inflation fixed lungs (n=3, bar=100 μm). (i) Overlay of CD11b vs. CD11c staining from BAL cells and blood neutrophils of day 3 LPS-challenged HckF/F mice (blue: BAL cells, red: blood neutrophils; n=3–5). (j–l) Flow cytometry used to calculate total AMΦ, monocyte, neutrophil, and lymphocyte numbers (n=3–5, *P<0.05, **P<0.01, two-tailed Student’s t-test: j and k, Welch t-test: l). (m, n) Mean fluorescence intensity (MFI) of AMΦ CD11b expression (total AMΦ CD11b MFI−non-binding antibody control AMΦ MFI) in Dulbecco’s modified Eagle’s medium (DMEM) control and day 3 LPS-challenged SHIP-1−/− and HckF/F mice (n=3–7, *P<0.05, Student’s t-test).
In vivo targeting of CD11bpos lung macrophages
CD11b is ubiquitously expressed on monocytes and neutrophils, which prevents the exclusive targeting of AMΦs using anti-CD11b mAbs. However, as CD11b is discretely expressed on inflammatory AMΦs in mice, we used anti-CD11b administration to test whether CD11bpos AMΦs could be selectively blocked without perturbations to homeostatic CD11bneg AMΦs. Intranasal anti-CD11b Ab administration 24 h post-LPS challenge attenuated lung inflammation by significantly decreasing lung neutrophil and monocyte infiltration in C57BL/6 mice 3 days post-LPS challenge (Figure 4a–d). Interestingly, AMΦ numbers remained unchanged following anti-CD11b Ab administration (Figure 4d), suggesting that anti-CD11b blocked surface AMΦ CD11b expression rather than depleting CD11bpos AMΦs. Anti-CD11b Ab administration demonstrated a dose-dependent preferential neutralization of CD11bhigh neutrophils before CD11bhigh lung monocytes and CD11bpos AMΦs (Figure 4e). Anti-CD11b also attenuated lung neutrophilia in a dose-dependent manner (Figure 4f,g). Decreased lung tissue apoptosis was observed in the lungs of anti-CD11b Ab–treated, but not vehicle-PBS or rat IgG isotype control Ab–treated C57BL/6 mice at day 3 post-LPS challenge (Figure 4h,i). In contrast, CD11b−/− mice did not show attenuated neutrophil or monocyte recruitment day 3 post-LPS challenge but did show variable AMΦ and monocyte CD11a expression and upregulated monocyte CD11c surface expression (see Supplementary Figure S5 online).
Anti-CD11b antibody blocks CD11bpos but not CD11blow/neg alveolar macrophages (AMΦs) post-lipopolysaccharide (post-LPS) challenge in C57BL/6 mice. (a) LPS-challenged C57BL/6 mice were administered rat IgG isotype control or anti-CD11b antibody (Ab) 24 h after LPS challenge. Hematoxylin and eosin–stained sections of inflation fixed lungs day 3 post-LPS challenge (n=4, scale=100 μm). (b, c) Flow cytometry used to calculate total AMΦ, monocyte, neutrophil, and lymphocyte numbers (n=6, *P<0.05, **P<0.01, ***P<0.005, analysis of variance (ANOVA) test with Tukey’s post-test). (d) CD11b vs. CD11c expression profile of bronchoalveolar lavage (BAL) cells at day 3 post-LPS challenge with anti-CD11b or isotype control Ab administration. (e) Dose-dependent neutrophil, monocyte, and CD11bpos AMΦ neutralization following anti-CD11b Ab administration (0.1, 0.5, or 1.0 μg g−1 mouse weight). (f, g) Flow cytometry used to calculate total AMΦ, monocyte, neutrophil, and lymphocyte numbers (n=3–7, *P<0.05, ANOVA test with Tukey’s post-test). (h) TUNEL (terminal deoxinucleotidyl transferase-mediated dUTP-fluorescein nick end labeling) staining used to measure total lung immune and non-immune cell apoptosis in day 3 post-LPS challenge treatment groups (n=3 with 3 sections of lung consolidations analyzed per mouse, *P<0.05, ANOVA test with Tukey’s post-test). (i) Representative sections of TUNEL and anti-CD45 Ab–stained lung tissue. DAPI (4,6-diamidino-2-phenylindole): blue, cleaved nuclei indicating cell apoptosis: green, CD45pos cells: red and cell membrane staining: purple, red arrows: TUNELpos immune cell, black arrows: TUNELpos non-immune cell, bar=30 μm). PMN, polymorphonuclear neutrophil.
Identification of analogous human immune cell profiles
As distinct BAL cell immunophenotypes were clearly discerned in mice, we investigated whether parallel immune cell features could be identified in humans. Blood neutrophil CD11b expression was similar between healthy volunteers and respiratory patients seen in the day clinic, although CD11b variation is higher in humans than in mice (Figure 5a). Nevertheless, >92% of all patient blood neutrophils existed within the same gated CD11b expression range used to reference AMΦ CD11b expression (see Supplementary Figure S6a,b online). AMΦ to blood neutrophil CD11b expression clearly distinguished between individual patients with lower or higher AMΦ CD11b expression analyzed on the same day independently of data acquisition variations (Figure 5b). AMΦ CD11b upregulation could also be confirmed by a comparison of AMΦ CD11b and CD14 expression levels and standard AMΦ CD11b MFI calculations (Figure 5c; MFI: 101 and 270 for CD11b expression on PT4912A and PT4912B, respectively).
A complementary approach for the characterization of human bronchoalveolar lavage (BAL) inflammatory profiles. (a) Histograms of human blood neutrophil CD11b expression; each of the three histograms depict n=2 random patient and 2 healthy volunteer blood samples (black: neutrophil Mac-1 expression, dotted gray: non-binding antibody (Ab) control). (b) Overlay of CD11b vs. CD33 staining of patient BAL cells and blood neutrophils (blue: BAL cells, red: blood neutrophils, boxed regions: alveolar macrophage (AMΦ) CD11b expression relative to the gated region of blood neutrophil CD11b expression). Samples collected and analyzed on the same day. (c) AMΦ CD11b and CD14 expression represented as histograms (black: CD11b, gray: CD14, dotted gray: non-binding Ab control). (d) CD11b vs. CD33 staining of patient BAL cells show the presence of a CD11bhighCD33lowCD15high subset, which correlates with the profile of eosinophils (R=residential AMΦs, E=eosinophils, blue: BAL cell staining, gray: non-binding Ab control). (e) Patient AMΦ and eosinophil BAL subsets further characterized by Class II (HLA-DR), CD14, and CD86 expression and represented as histograms (dotted gray: non-binding Ab control). (f) Patient BAL cytospins stained using Hemacolor (Merck Millipore); eosinophils appear as polymorphonuclear leukocytes with pink-red cytoplasmic staining (white arrows; bar=20 μm). MFI, mean fluorescence intensity; PMN, polymorphonuclear neutrophil.
For human BAL cell immunophenotyping, CD33 was chosen over CD11c as CD33 is a commonly used pan MΦ marker in humans,28 which separated AMΦs more clearly than CD11c in our studies and thus is a more definitive marker of human AMΦs (see Supplementary Figure S7a online). As human lung monocytes are CD33posCD11bhigh and potentially similar to CD33posCD11bhigh AMΦs, monocytes were excluded from our gating strategy based on their lack of autofluorescence and smaller size (low forward scatter (FSC)) to eliminate signal contamination (see Supplementary Figure S7b online). Despite this, an additional CD33lowCD11bhigh immune cell subset was still prominent in the BAL of a subset of human patients (Figure 5d, subset E). This subset showed negligible Class II, CD14, or CD86 expression (Figure 5e) and was identified as lung eosinophils from cell differentials of BAL cytospins (Figure 5f; granular cells with eosinophilic cytoplasm as denoted by white arrows). This allowed the inclusion of lung eosinophilia as a parameter for immunophenotyping, similar to mice. The degree of lung neutrophilia, another preestablished parameter for COPD and asthma patient immunophenotyping, could simultaneously be measured as the proportion of granular CD15highCD11bhigh cells in BAL(see Supplementary Figure S1 online and Figure 6a–d, pie charts).
Identification of subgroups of humans with shared inflammatory bronchoalveolar lavage (BAL) profiles. (a–d) CD11b vs. CD33 staining of patient BAL cells (alveolar macrophages (AMΦs)±eosinophils or other atypical myeloid subsets as featured in PT5012, blue: BAL cells, red: blood neutrophils, left panels). Histograms represent AMΦ CD11b and CD14 expression with respect to non-binding antibody (Ab) control (black: CD11b expression, gray; CD14 expression, dotted gray: non-binding Ab control, middle panels). Total BAL cell differentials calculated using flow cytometry and presented as a percentage of total live leukocytes (AMΦs: orange, polymorphonuclear neutrophils (PMNs): dark blue, other innate immune subsets: gray, CD4+ T cells: red, CD8+ T cells: green, other lymphocytes: purple, right panels). (e) Total distribution of the mean fluorescence intensity (MFI) of AMΦ CD11b and CD14 expression in all analyzed patients (n=14 patients).
Our immunophenotyping approach could thus incorporate AMΦ CD11b expression alterations with other preexisting parameters for clinical endotyping. Using these parameters, we analyzed human BAL from a randomly sampled pool of patients undergoing bronchoscopies for clinical diagnosis (see Supplementary Table S3 online). Human patients could be broadly segregated into one of four lung immunophenotypes: into patients with (a) only CD11blow AMΦs, (b) only CD11bmod AMΦs, (c) only CD11bhigh AMΦs, and (d) CD11bmod/high AMΦs with eosinophilia or atypical myeloid cell infiltration (Figure 6a–d). Surprisingly, this mirrored the distinct immunophenotypes observed in C57BL/6 compared with SHIP-1−/− and HckF/F mice. Patients with CD11blow AMΦs (set as having ⩽10% AMΦs with higher CD11b expression than blood neutrophils) showed low neutrophil and monocyte infiltration and most closely resembled the BAL phenotype of healthy C57BL/6 mice (Figure 6a). Patients with CD11bmod AMΦs were arbitrarily characterized to serve as an intermediate subgroup (with 10–40% CD11bpos AMΦs with higher CD11b expression than blood neutrophils) to distinguish between patients with obvious CD11blow AMΦ or CD11bhigh AMΦ phenotypes (Figure 6b). Patients with CD11bhigh AMΦs (⩾40% AMΦs with higher CD11b expression than blood neutrophils) but negligible monocyte or eosinophil recruitment greatly resembled the immunophenotype of SHIP-1−/− mice and showed increased CD4+ and CD8+ T-cell proportions in BAL (Figure 6c). Patients with CD11bmod/high AMΦs and lung eosinophilia resembled the BAL cell immunophenotype of HckF/F mice or C57BL/6 mice during the clearance phase of influenza infection (Figure 6d). Interestingly, a unique subset of CD33highCD11blow non-lymphocyte cells was additionally found in the BAL of PT5012 (Figure 6d). These atypical myeloid cells were lobe-specific to disease pathology (see Supplementary Figure S8 online).
As human AMΦ CD11b expression exists in a continuum, larger-scale clinical studies are required to identify CD11bhigh AMΦ cutoffs, which correlate with increased lung or systematic inflammation and disease burden. Our results demonstrate that patients with CD11blow or CD11bhigh AMΦs exist and can be identified in a directly comparable manner. To represent AMΦ CD11b expression in an unbiased manner, we additionally calculated AMΦ CD11b MFIs and AMΦ to blood neutrophil CD11b MFI ratios for each patient (Figure 6a–d and Supplementary Table S2 online). MFI values fluctuate between individual experiments even with the usage of single peak fluorescent beads. However, patients categorized into the subgroups A and C were clearly distinguished by all calculation parameters (see Supplementary Table S2 online). As additional confirmation of CD11b selective AMΦ expression heterogeneity, we simultaneously measured AMΦ CD14 expression in the same experiment. In contrast to CD11b, CD14 AMΦ surface expression remained relatively constant between individual patients independently of AMΦ CD11b expression differences or lung disease status (Figure 6a–d). Moreover, while blood neutrophil CD11b expression was relatively constant between patients, blood vs. lung neutrophil CD11b upregulation was observed in LPS-challenged C57BL/6 mice, SHIP-1−/− and HckF/F mice and human patients with similar inflammatory BAL cell profiles (see Supplementary Figure S9a,b online). The degree of blood vs. lung neutrophil CD11b upregulation positively correlated with AMΦ CD11b upregulation (r=0.806) and further distinguished between patients with CD11blow or CD11bmod/high AMΦs, providing an additional parameter of local lung inflammation (see Supplementary Figure S9b online).
Discussion
Our study describes a simplified and internally standardized flow cytometry approach for human lung immunophenotyping, adapted from the study of inflammatory lung conditions in mice. Critically, it is the first approach that can systematically profile AMΦ-based alterations alongside preexisting immune cell parameters in a directly comparable manner. AMΦs are the predominant immune cell population within the lung parenchyma and express an expansive repertoire of cell surface molecules, which dynamically reorganize in response to different environmental stimuli.29 Theoretically, this should render them as more selective reporters of the inflammatory microenvironment compared with current parameters such as lung neutrophilia or eosinophilia, which only indicate immune subset presence and not phenotype.
In truth, however, there are very few disease-associated MΦ markers in humans or even mice. The first reason stems from their dynamic and expansive nature; MΦs activate a spectrum of transcriptional programs in response to a diverse array of environmental stimuli in a time-dependent manner.30 This creates confusion in the absence of common descriptive standards as different studies ascribing markers to different MΦ activation states may not actually match each other.5 For instance, induced nitric oxide synthase and Arginase I expression have been extensively used to discriminate between more classically M(LPS or interferon-γ) or alternatively M(interleukin-4) activated MΦs yet their presence is confounded by mouse strain–specific differences in enzyme expression and species-dependent differences in arginine and nitric oxide generation.31, 32 Recently, transcriptome studies of tissue MΦ and dendritic cell subsets and differentially stimulated human monocyte-derived MΦs have identified new genes with selective expression in different tissue MΦ subsets or activation stimuli (such as vascular cell adhesion molecule 1 for splenic MΦs, intercellular adhesion molecule-2 for peritoneal MΦs, Siglec H for microglial).30, 33, 34, 35 This unbiased and systematic approach may generate new disease-specific MΦ markers for further evaluation and renders the discovery of systematic human MΦ profiling methods an even more critical objective.
A second obstacle lies in the difficulty of translating candidate MΦ markers from animal or in vitro studies. Few surface markers are similarly expressed on mouse and human AMΦs. Translatable MΦ activation markers are so far limited to several chemokines, scavenger receptors, and matrix proteins.5 As the only cell surface receptors from the list, scavenger receptors are the most pragmatic candidates for immunophenotyping. However, apart from MARCO,36 few scavenger receptors are even dually expressed on mouse and human AMΦs, let alone in parallel stimuli-selective patterns. More importantly, the expression of these and many other classes of MΦ surface proteins is predominantly continuous.37, 38 Continuously expressed markers are represented as MFIs, which permits semi-quantitative comparisons within single flow cytometry experiments but hinders comparisons and data replication between independent studies. MFIs also do not provide information about cell population spread, may be altered by different gating strategies, and can be inadvertently skewed by extremely low or high value false events captured, future questioning their usefulness as meaningful clinical readouts. Thus, without alternative study strategies, many future candidate human MΦ markers may be rejected in the screening process in favor of binary observations, which are biologically unlikely. A conventional myeloid cell parameter would be the frequency of CD14posCD16neg pro-inflammatory monocytes, CD14posCD16pos vs. CD14lowCD16pos patrolling monocytes in individual patients,39 which can be used to describe the local environment in terms of recruited monocyte phenotypes. However, residential MΦs possess different and often opposing functions to recruited monocytes and an understanding of residential MΦ responses may provide a more accurate reflection of the local inflammatory state. Using our flow cytometry approach, we show that AMΦ CD11b upregulation occurs in both mice and human patients alongside other inflammatory changes despite basal CD11b expression on human AMΦs. In particular, our studies suggest that CD11bhigh AMΦs may identify an important subset of patients with inflammatory lung conditions and that a combinational immunophenotyping approach that monitors AMΦ CD11b expression alongside the absence or presence of other innate immune cell subsets may provide a more personalized understanding of individual patient lung microenvironments.
CD11b is a leukocyte integrin composed of the CD11b α subunit and common integrin β2 subunit and binds to ligands intercellular adhesion molecule-1, iC3b (cleaved product of complement 3b), and fibrinogen.40 CD11b function was first suggested when a deficiency in the common integrin β2 subunit (and hence LFA-1, CD11b, and p150/95 assembly and surface expression) was identified in a group of patients with recurrent bacterial infections owing to defective neutrophil and myeloid cell adhesion and phagocytosis.41 Later studies identified CD11b upregulation as a functional component of neutrophil aggregation but not neutrophil endothelial cell adhesion or extravasation.42 Surprisingly, global CD11b deletion in mice does not induce prominent physiological alterations or attenuate neutrophil accumulation in inflamed tissues.43, 44 In CD11b−/− mice, however, complement-mediated phagocytosis and vasculitis are significantly attenuated,43, 44 most likely through a failure of iC3b ligand–mediated activation of CD11b receptor signaling. From our studies, given that anti-CD11b Ab administration but not CD11b deletion attenuated lung inflammation in mice, global CD11b deletion may not replicate the consequences of transiently targeting CD11b in chronic diseases. As blocking of surface AMΦ CD11b rather than depletion of CD11bpos AMΦs most likely occurred following anti-CD11b Ab administration, it is crucial to dissect whether AMΦ CD11b signaling itself or CD11bpos/high AMΦs are potential effectors of acute and chronic lung inflammation. Specifically, the comparison of CD11bneg (or CD11blow in humans) and CD11bpos/high AMΦ transcriptomes may identify whether alterations in CD11b-related signaling pathways or a broader array of effector pathways exist between these two AMΦ subpopulations.
AMΦ CD11b expression alterations have been previously described in clinical studies of acute and chronic inflammatory lung diseases. CD11bhigh AMΦs correlated with increased lung monocyte influx and respiratory failure severity in a 21-day follow-up of ARDS patients requiring mechanical ventilation.45 AMΦ CD11b upregulation was also observed in patients with pneumonia compared with healthy volunteers in a separate study that correlated CD11b AMΦ expression with the level of neutrophil chemokine interleukin-8.46 Interestingly, AMΦ CD11b expression was not significantly different between COPD patients and healthy non-smokers in a previous study, though this may reflect a recruitment of inflammatory vs. non-inflammatory COPD endotypes.47 Overall, the current diversity of CD11b AMΦ– and lung eosinophil–associated mouse models of emphysema temptingly suggests that distinct lung innate immune cell niches may be present in different COPD endotypes. Lung immunophenotyping in previously established COPD animal models may also help the field to more clearly differentiate immune- vs. non-immune-driven mechanisms of human COPD pathogenesis.48
Although our approach can be immediately used in any flow cytometer facility, small caveats remain that apply to all lung immunophenotyping studies. Sample storage needs to be consistent for large-scale usage.49 We used freshly obtained BAL samples for our studies in order to match the in vivo immunophenotype as closely as possible. This may be difficult in places where same-day flow cytometry analysis is not possible and cryopreservation options have to be explored. Cigarette smoke–induced cell autofluorescence is also a major obstacle as background AMΦ signals are exponentially increased in current smokers, confounding fluorescence AMΦ read-outs. Current cigarette smokers (<6 months) were excluded from our study, but this may be undesirable for COPD endotyping. In unavoidable scenarios, it is possible that non-fluorescence detection methods such as mass cytometry may be employed to characterize all samples from a similar baseline, although non-specific Ab-binding issues may still exist.
In summary, our study introduces a new internally standardized and hence directly comparable approach for AMΦ immunophenotyping. Used in combination with other established parameters for clinical endotyping (the presence of eosinophilia or neutrophilia), we show that inflammation-associated CD11b AMΦ profiles exist heterogeneously in both mouse lung disease models and human patients. This is timely as our understanding of immune cell contributions to disease pathogenesis and disease endotyping advances and the technology to discriminate differential immune cell subsets becomes more widely accessible.
Methods
Study approval
Recruited patients undergoing a bronchoscopy for clinical diagnosis participated voluntarily and provided informed consent under protocols approved by the Melbourne Health Institutional review board. Written informed consent was obtained from all subjects prior to inclusion.
Human study subjects
Non-targeted or peri-lesion BAL were performed depending on computed tomographic scan presentations using 3 × 20 ml saline, and patient BAL cells were collected for fluorescence-activated cell sorting (FACS) analysis. Three milliliters of blood was also collected from each patient. Patient diagnosis was established by same-day tissue biopsy (see Supplementary Table S3 online). A total of 14 patient samples were analyzed with standardized machine settings using fluorescent beads (allophycocyanin (APC) and APC-Cy7 fluorescence reference standards, Bangs Laboratories, Fishers, IN). Patients with BAL samples showing highly autofluorescent AMΦs owing to recent cigarette smoke exposure were excluded from analysis as elevated autofluorescence greatly masks the surface expression of markers in AMΦs.
Mice
SHIP-1−/−16 and HckF/F22 mice have been described and were crossed onto the C57BL/6 background for ⩾7 generations. C57BL/6 CD11b−/− mice were a gift from Professor Karlheinz Peter (Baker IDI Heart and Diabetes Institute, Melbourne, Australia). C57BL/6 mice were purchased from the Monash Animal Services (ARL, Melbourne, Australia). All mice were kept in specific pathogen-free conditions (Monash MARP and MICU, Melbourne, Australia). All animal experiments were approved by the Alfred Medical Research and Education Precinct (AMREP) Animal Ethics Committee in accordance with National Health and Medical Research Council (NHMRC) Australia guidelines.
LPS challenge
Eight–12-week-old C57BL/6, CD11b−/−, SHIP-1−/−, or HckF/F mice were lightly anesthetized with 5% isofluorane and intranasally administered 30 μl of Escherichia coli LPS (10 μg per mouse, serotype O114:B4; Sigma, Saint Louis, MO). LPS-challenged mice were euthanized at day 3 post-LPS challenge. Vehicle control C57BL/6 mice received 30 μl of Dulbecco’s modified Eagle’s medium.
Mem71 infection
Eight–12-week-old C57BL/6 mice were lightly anesthetized and intranasally delivered 30 μl Mem71 influenza virus (31,000 pfu per mouse, sublethal dose). Mice were euthanized at day 10 postinfection.
Antibody treatment
Twenty-four hours post-LPS challenge, C57BL/6 mice were lightly anesthetized and intranasally administered 30 μl of anti-CD11b antibody (1.0, 0.5 or 0.1 mg kg−1, clone M1/70, Bio X cell, West Lebanon, NH) or rat IgG isotype control (1 mg kg−1, clone LTF-2, Bio X cell). Mice were euthanized on day 3. Ab administration occurred at 24 h post-LPS as this marks the onset of AMΦ Mac-1 upregulation15 and minimizes the neutralization of CD11bhigh lung neutrophils recruited earlier.
Flow cytometry
For mouse BAL, filtered BAL and red blood cell–lysed blood cells were resuspended in FACS buffer (Mouse PBS, 2% fetal calf serum, 2 mM EDTA) and incubated with Fc block (0.6 mg ml−1, anti-mouse CD32/CD16, clone 2.4G2, 10 min). Cells were stained with 1° monoclonal Abs or non-binding Ab controls (20 min, 4 °C on ice, listed in Supplementary Table S2 online) followed by 2° fluorophore conjugates where necessary (streptavidin APC-A750, 15 min, 4 °C on ice). Cells were then washed and resuspended in 50 μl FACS buffer with 5 μl DAPI (4,6-diamidino-2-phenylindole) solution (1 μg ml−1, Sigma Aldrich, Saint Louis, MO) before analysis. Our FACS strategy has been previously described.15 To examine mouse AMΦs (see Supplementary Figure S1a online), DAPIneg cells were first gated to exclude all dead cells from analysis. Anti-mouse CD45-fluorescein isothiocyanate was next used to positively select all BAL leukocytes. Gating of FSC-A vs. FSC-W cells excluded doublet cells. Ly6g-phycoerythrin was then used to negatively gate out all neutrophils, and all lymphocytes were gated out based on their lower FSC-A and SSC-A profile. This allowed FACS analysis on the remaining MΦ/monocyte subpopulations. AMΦs were discriminated from dendritic cells based on their high autofluorescence in an unstained PerCPCy5.5 channel.
For human BAL (all samples in Figures 5 and 6), filtered BAL and red blood cell–lysed blood cells were resuspended in FACS buffer (Human PBS, 2% fetal calf serum, 2 mM EDTA) and incubated with saturated Mouse IgG to block non-specific 1° antibody binding to human cells (2.5 μg per 250,000 cells, Normal mouse IgG, Merck Millipore, Billerica, MA, 20 min on ice). Stains contained 250,000 cells per tube. Cells were stained with 1° monoclonal Abs or non-binding Ab controls (20 min, 4 °C on ice; listed in Supplementary Table S5 online), washed, and resuspended in 100 μl FACS buffer, with 5 μl DAPI solution (1 μg ml−1, Sigma Aldrich) added before analysis. Critically, prior to each experiment, single-peak fluorescent calibration beads were used to maintain a standardized laser voltage for data acquisition (APC and APC-Cy7 fluorescence reference standards, Bangs Laboratories). To examine human AMΦs (see Supplementary Figure S1b online), DAPIneg cells were first positively selected to exclude all dead cells from analysis. Live CD45pos singlet cells were positively selected and neutrophils were negatively gated by CD15high staining with low FSC. AMΦs and eosinophils are positively selected by CD15high cells with higher FSC (eosinophils are CD15high and AMΦs are autofluorescent in phycoerythrin). For AMΦ MFI calculations, events are further gated in the PerCP5.5 channel for only autofluorescent AMΦs. The samples used to generate the preliminary human data shown in Figure 1a were incubated with Human TruStain FcX Fc block (Biolegend, San Diego, CA; 5 μl per 250,000 cells, 20 min on ice) instead of purified mouse IgG.
All FACS data was acquired on a LSR-Fortessa (BD Biosciences, San Jose, CA) except Supplementary Figure S2 (LSR-Canto II, BD Biosciences), and ⩾5,000 AMΦ events per sample was collected. Analysis was performed using the FlowJo software (Mac V8.7.3 or Windows V10, FlowJo LLC, Ashland, OR).
Statistics
All values are expressed as mean±s.e.m. from at least two independent experiments. Unpaired two-tailed Student’s t-test was performed for all studies comparing two sets of data, except where the s.d. of data sets differed and a Welch t-test was used. Studies comparing three sets of data were analyzed by the analysis of variance test followed by the Tukey’s post-test for statistical significance. The correlation between human AMΦ CD11b and CD86 surface expression was determined by linear regression. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software Inc, La Jolla, CA).
References
Betts, M.R. et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107, 4781–4789 (2006).
Ghia, P. et al. The pattern of CD38 expression defines a distinct subset of chronic lymphocytic leukemia (CLL) patients at risk of disease progression. Blood 101, 1262–1269 (2003).
Kok, A. et al. Early initiation of combined antiretroviral therapy preserves immune function in the gut of HIV-infected patients. Mucosal. Immunol. 8, 127–140 (2015).
Geissmann, F., Jung, S. & Littman, D.R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 19, 71–82 (2003).
Murray, P.J. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 41, 14–20 (2014).
Lotvall, J. et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J. Allergy. Clin. Immunol. 127, 355–360 (2011).
Moore, W.C. et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am. J. Respir. Crit. Care Med. 181, 315–323 (2010).
Thomsen, M., Dahl, M., Lange, P., Vestbo, J. & Nordestgaard, B.G. Inflammatory biomarkers and comorbidities in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 186, 982–988 (2012).
Agusti, A. et al. Persistent systemic inflammation is associated with poor clinical outcomes in COPD: a novel phenotype. PLoS One 7, e37483 (2012).
Hurst, J.R. et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N. Engl. J. Med. 363, 1128–1138 (2010).
Kasahara, Y., Tuder, R.M., Cool, C.D., Lynch, D.A., Flores, S.C. & Voelkel, N.F. Endothelial cell death and decreased expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in emphysema. Am. J. Respir. Crit. Care Med. 163, 737–744 (2001).
Morris, D.G. et al. Loss of integrin alpha(v)beta6-mediated TGF-beta activation causes Mmp12-dependent emphysema. Nature 422, 169–173 (2003).
Shibata, Y., Zsengeller, Z., Otake, K., Palaniyar, N. & Trapnell, B.C. Alveolar macrophage deficiency in osteopetrotic mice deficient in macrophage colony-stimulating factor is spontaneously corrected with age and associated with matrix metalloproteinase expression and emphysema. Blood 98, 2845–2852 (2001).
Oh, S.Y. et al. Src homology 2 domain-containing inositol 5-phosphatase 1 deficiency leads to a spontaneous allergic inflammation in the murine lung. J. Allergy Clin. Immunol. 119, 123–131 (2007).
Duan, M., Li, W.C., Vlahos, R., Maxwell, M.J., Anderson, G.P. & Hibbs, M.L. Distinct macrophage subpopulations characterize acute infection and chronic inflammatory lung disease. J. Immunol. 189, 946–955 (2012).
Helgason, C.D. et al. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 12, 1610–1620 (1998).
Rauh, M.J. et al. SHIP represses the generation of alternatively activated macrophages. Immunity 23, 361–374 (2005).
Harder, K.W. et al. Perturbed myelo/erythropoiesis in Lyn-deficient mice is similar to that in mice lacking the inhibitory phosphatases SHP-1 and SHIP-1. Blood 104, 3901–3910 (2004).
Takeshita, S. et al. SHIP-deficient mice are severely osteoporotic due to increased numbers of hyper-resorptive osteoclasts. Nat. Med. 8, 943–949 (2002).
Maxwell, M.J., Duan, M., Armes, J.E., Anderson, G.P., Tarlinton, D.M. & Hibbs, M.L. Genetic segregation of inflammatory lung disease and autoimmune disease severity in SHIP-1-/- mice. J. Immunol. 186, 7164–7175 (2011).
Sly, L.M., Rauh, M.J., Kalesnikoff, J., Song, C.H. & Krystal, G. LPS-induced upregulation of SHIP is essential for endotoxin tolerance. Immunity 21, 227–239 (2004).
Ernst, M. et al. Constitutive activation of the SRC family kinase Hck results in spontaneous pulmonary inflammation and an enhanced innate immune response. J. Exp. Med. 196, 589–604 (2002).
Johnston, L.K., Rims, C.R., Gill, S.E., McGuire, J.K. & Manicone, A.M. Pulmonary macrophage subpopulations in the induction and resolution of acute lung injury. Am. J. Respir. Cell Mol. Biol. 47, 417–426 (2012).
Stevens, W.W., Kim, T.S., Pujanauski, L.M., Hao, X. & Braciale, T.J. Detection and quantitation of eosinophils in the murine respiratory tract by flow cytometry. J. Immunol. Methods 327, 63–74 (2007).
Mallia, P. et al. Experimental rhinovirus infection as a human model of chronic obstructive pulmonary disease exacerbation. Am. J. Respir. Crit. Care Med. 183, 734–742 (2011).
Aaron, S.D. et al. Granulocyte inflammatory markers and airway infection during acute exacerbation of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 163, 349–355 (2001).
Bafadhel, M. et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am. J. Respir. Crit. Care Med. 184, 662–671 (2011).
Tanimoto, M., Scheinberg, D.A., Cordon-Cardo, C., Huie, D., Clarkson, B.D. & Old, L.J. Restricted expression of an early myeloid and monocytic cell surface antigen defined by monoclonal antibody M195. Leukemia 3, 339–348 (1989).
Hussell, T. & Bell, T.J. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 14, 81–93 (2014).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 (2014).
Sans-Fons, M.G. et al. Arginine transport is impaired in C57Bl/6 mouse macrophages as a result of a deletion in the promoter of Slc7a2 (CAT2), and susceptibility to Leishmania infection is reduced. J. Infect. Dis. 207, 1684–1693 (2013).
Schneemann, M. & Schoedon, G. Species differences in macrophage NO production are important. Nat. Immunol. 3, 102–102 (2002).
Gautier, E.L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Chiu, I.M. et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 4, 385–401 (2013).
Dutta, P. et al. Macrophages retain hematopoietic stem cells in the spleen via VCAM-1. J. Exp. Med. 212, 497–512 (2015).
Palecanda, A. et al. Role of the scavenger receptor MARCO in alveolar macrophage binding of unopsonized environmental particles. J. Exp. Med. 189, 1497–1506 (1999).
Misharin, A.V., Morales-Nebreda, L., Mutlu, G.M., Budinger, G.R. & Perlman, H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol. 49, 503–510 (2013).
Kaku, Y. et al. Overexpression of CD163, CD204 and CD206 on alveolar macrophages in the lungs of patients with severe chronic obstructive pulmonary disease. PLoS One 9, e87400 (2014).
Oshansky, C.M. et al. Mucosal immune responses predict clinical outcomes during influenza infection independently of age and viral load. Am. J. Respir. Crit. Care Med. 189, 449–462 (2014).
Corbi, A.L., Kishimoto, T.K., Miller, L.J. & Springer, T.A. The human leukocyte adhesion glycoprotein Mac-1 (complement receptor type 3, CD11b) alpha subunit. Cloning, primary structure, and relation to the integrins, von Willebrand factor and factor B. J. Biol. Chem. 263, 12403–12411 (1988).
Anderson, D.C. & Springer, T.A. Leukocyte adhesion deficiency: an inherited defect in the Mac-1, LFA-1, and p150,95 glycoproteins. Annu. Rev. Med. 38, 175–194 (1987).
Vedder, N.B. & Harlan, J.M. Increased surface expression of CD11b/CD18 (Mac-1) is not required for stimulated neutrophil adherence to cultured endothelium. J. Clin. Invest. 81, 676–682 (1988).
Lu, H. et al. LFA-1 is sufficient in mediating neutrophil emigration in Mac-1-deficient mice. J. Clin. Invest. 99, 1340–1350 (1997).
Hirahashi, J. et al. Mac-1 signaling via Src-family and Syk kinases results in elastase-dependent thrombohemorrhagic vasculopathy. Immunity 25, 271–283 (2006).
Rosseau, S. et al. Phenotypic characterization of alveolar monocyte recruitment in acute respiratory distress syndrome. Am. J. Physiol. Lung Cell. Mol. Physiol. 279, L25–L35 (2000).
Buhling, F. et al. MRP8/MRP14, CD11b and HLA-DR expression of alveolar macrophages in pneumonia. Immunol. Lett. 71, 185–190 (2000).
Lofdahl, J.M., Wahlstrom, J. & Skold, C.M. Different inflammatory cell pattern and macrophage phenotype in chronic obstructive pulmonary disease patients, smokers and non-smokers. Clin. Exp. Immunol. 145, 428–437 (2006).
Ueno, M. et al. Alendronate inhalation ameliorates elastase-induced pulmonary emphysema in mice by induction of apoptosis of alveolar macrophages. Nat. Commun. 6, 6332 (2015).
Maecker, H.T., McCoy, J.P. & Nussenblatt, R. Standardizing immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol. 12, 191–200 (2012).
Acknowledgements
We thank Karlheinz Peter for providing CD11b−/− mice; the respiratory registrars and Michelle Thomson at the Royal Melbourne Hospital for patient consent; Maverick Lau for TUNEL staining protocols; Evelyn Tsantikos for SHIP-1−/− genotyping; the AMREP flow cytometry facility for support; Stephen Cody and Chad Johnson from Monash Micro Imaging (Monash University) for scientific and technical assistance; and Kylie Spark, Steve Comber; and Naomi Borg from MICU and MARP animal facilities (Monash University) for monitoring the animals. This work was made possible through funding from the NHMRC, Australia; project grants 1008298 and 1080274 (to M.L.H. and G.P.A.) and 1067244 (to M.E.), program grant 1071916 (to W.C.) and support from the Victorian State Government Operational Infrastructure Scheme. M.D. was a recipient of a Cancer Research Institute Scholarship and the Nick Christopher Top-Up Scholarship, while M.E., W.C. and M.L.H. were supported by NHMRC Research Fellowships.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Additional information
Author contribution
M.D. designed experiments, performed all mouse and human studies, analyzed and interpreted all data, conceptualized the study, and wrote the manuscript. D.P.S., D.S., M.H., and L.B.I. performed lung lavages, provided patient samples, and interpreted clinical data. W.C. interpreted data and edited the manuscript. M.E. provided a critical reagent and edited the manuscript. G.P.A. was involved in conceptualizing and funding the study and edited the manuscript. M.L.H. designed experiments, conceptualized and funded the study, interpreted all data, and edited the manuscript. All authors approved the final draft.
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Supplementary information
Rights and permissions
About this article
Cite this article
Duan, M., Steinfort, D., Smallwood, D. et al. CD11b immunophenotyping identifies inflammatory profiles in the mouse and human lungs. Mucosal Immunol 9, 550–563 (2016). https://doi.org/10.1038/mi.2015.84
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2015.84
This article is cited by
-
Inhibition of tyrosine kinase Fgr prevents radiation-induced pulmonary fibrosis (RIPF)
Cell Death Discovery (2023)
-
Mesenchymal stem cells exert their anti-asthmatic effects through macrophage modulation in a murine chronic asthma model
Scientific Reports (2022)
-
Intratracheal administration of mesenchymal stem cells modulates lung macrophage polarization and exerts anti-asthmatic effects
Scientific Reports (2022)
-
A multistep in vitro hemocompatibility testing protocol recapitulating the foreign body reaction to nanocarriers
Drug Delivery and Translational Research (2022)
-
Phosphatidylserine externalized on the colonic capillaries as a novel pharmacological target for IBD therapy
Signal Transduction and Targeted Therapy (2021)