Abstract
Interleukin (IL)-17C is a novel member of the IL-17 cytokine family. Its function in human inflammatory bowel disease (IBD) remains elusive as its role in colonic inflammation is entirely derived from murine models. We aimed to analyze the role of IL-17C in human IBD, focusing on T helper type 17 (Th17) cell- and intestinal epithelial cell (IEC)-dependent mechanisms. IL-17C mRNA (P=0.005), serum levels (P=0.008), and colonic staining intensity (P=0.004) is increased in active IBD. Serum IL-17C levels are modified by IL23R genotypes and IL-17C mRNA correlates (r>0.5, P<0.001) with IL-17A, tumor necrosis factor (TNF)-α, C-C motif chemokine ligand 20 (CCL20) and IL-23 mRNA in the inflamed colon of IBD patients. In the inflamed colon, IL-17C is produced by enteroendocrine and goblet cells, with contrary polar cytosolic localization of IL-17C within the cellular axis. In these two cell types, IL-17A strongly amplifies TNF-α-induced IL-17C production. On the molecular level, IL-17C production in IECs is dependent on TNF-α-activated nuclear factor-κB, extracellular signal–regulated kinase-1/2 and p38, and IL-17A-activated Akt, monocyte chemotactic protein–induced protein 1, and C/EBPδ. IL-17C upregulates the Th17 chemoattractant CCL20 in IECs. In summary, our findings support the involvement of IL-17A-amplified IL-17C production by enteroendocrine and goblet cells in the pathogenesis of active IBD, revealing an interaction between the neuroendocrine system and the Th17 pathway in human IBD.
Similar content being viewed by others
INTRODUCTION
Recently, numerous studies suggested an emerging role of T helper type 17 (Th17) cells in the pathogenesis of inflammatory bowel disease (IBD) (reviewed in refs. 1,2,3,4), supported by findings that many IBD susceptibility loci contain genes encoding proteins involved in Th17 cell differentiation.5, 6, 7, 8
Th17 cells are named after the hallmark cyokine IL-17A (IL-17), which is currently the best characterized cytokine of the IL-17 family. It is mainly produced by Th17 cells and also by innate T-cell subsets, such as natural killer T cells9 and γδ T cells.10 The main target cells of IL-17A are endothelial, epithelial, and stromal cells, in which IL-17A can elicit the release of pro-inflammatory molecules, mucins, and defensins (reviewed in Brand,1 Miossec and Kolls,11 and Gaffen12). Recently, a role of another IL-17 family member, IL-17C, in colonic inflammation has been proposed based on the observations that IL-17C is increased in dextran sodium sulfate (DSS)–induced murine colitis and that DSS sulfate–treated IL-17C knock-out mice and mice deficient of its receptor IL-17RE show exacerbated intestinal inflammation.13, 14, 15 Whereas the IL-17A receptor subunits IL-17RA and IL-17RC are broadly expressed in intestinal tissue, the mRNA expression of the IL-17C-specific receptor subunit IL-17RE is more limited, with the highest levels of IL-17RE mRNA to be found in murine colonic epithelial cells13, 14 and murine Th17 cells.16 In contrast to IL-17A, IL-17C is mainly produced by epithelial cells upon bacterial challenge and stimulation with cytokines.13, 14, 16, 17, 18 With regard to IBD, the induction of IL-17C by bacterial stimulation, e.g., via Toll-like receptor 5 signaling,17 is particularly interesting, as Toll-like receptors on intestinal epithelial cells (IECs) have been shown to be differentially expressed in IBD patients vs. healthy subjects.19, 20
Recently, a study by Hueber et al.21 reported unexpected detrimental effects of IL-17A neutralization by an anti-IL-17A antibody in patients with CD, despite beneficial effects of the same antibody in other Th17-mediated diseases, such as psoriasis, rheumatoid arthritis, and uveitis.22 Numerous murine models of intestinal inflammation, such as dextran sodium sulfate or trinitrobenzene sulfonic acid colitis, IKK-γ- and IL-10-dependent IBD or T-cell transfer–induced IBD, have provided valuable mechanistic insights in the pathogenesis of murine and human IBD.23, 24, 25, 26, 27 However, strong discrepancies between mouse models and human pathology in inflammatory diseases have been described very recently,28 as well as many differences between human and mouse immunology (e.g., Mestas and Hughes29). Importantly, the adverse effects of IL-17A neutralization in IBD patients have shown that therapeutic effects seen in murine models can often not be conferred to human settings, and—even more discouraging—that the beneficial effect of a specific antibody in several inflammatory diseases (e.g., psoriasis) can turn into detrimental effects of the same antibody in a related inflammatory disease, such as Crohn’s disease (CD).21 Therefore the involvement of a putative inflammatory mediator, such as IL-17C, in disease pathogenesis requires careful and distinct investigation depending on the affected tissue, emphasized by contrasting reports that suggest a pro-inflammatory role of IL-17C/IL-17RE in experimental psoriasis and encephalomyelitis but a protective role in experimental murine colitis.13, 14, 15, 16, 30 As the existing studies are entirely based on murine models of intestinal inflammation, we aimed to perform the first comprehensive analysis of the role of IL-17C in human IBD.
RESULTS
IL-17C mRNA and serum levels are elevated in IBD
In order to elucidate a potential role of IL-17C in IBD, we first analyzed its protein and mRNA levels in IBD patients and healthy subjects. Enzyme-linked immunosorbent assay (ELISA) detected significantly increased IL-17C levels in the serum of ulcerative colitis (UC) patients (43.8 pg ml−1 (28.1–59.6), n=61), compared with IL-17C serum levels of healthy controls (20.9 pg ml−1 (15.4–26.5), n=47) (P=0.008; Figure 1a). The increase in CD patients (25.3 pg ml−1 (14.4–26.2), n=64) was less pronounced and did not reach statistical significance. IL-17C protein was also detected in the serum of some healthy control patients. These results were confirmed using a second ELISA kit from another manufacturer utilizing different antibodies (Omnikine ELISA, Assay Biotech, Sunnyvale, CA; not shown).
Interleukin (IL)-17C serum protein and IL-17C mRNA levels in inflammatory bowel disease (IBD) patients and IL23R genotype correlations. (a) IL-17C is increased in the serum of ulcerative colitis (UC) patients, compared with healthy subjects; note that over-lying data points are a result of the threshold set to the minimal detectable IL-17C concentration in the enzyme-linked immunosorbent assay, previously acquired by spiking pooled human serum with rh-IL-17C. (b–d) IL17C mRNA levels in inflamed (n=20) and not-inflamed (n=48) colonic biopsies of IBD (UC, Crohn’s disease (CD)) patients. Arbitrary mRNA values were calculated manually using standard curve method, with normalization to the median mRNA values of HPRT1, YWHAZ, and RPL13A. Horizontal bars indicate the means of the respective values. (e) The mean level of IL-17C is decreased in UC patients (n=38) carrying one or two minor allele(s) (n=23) of the risk-decreasing IL23R single-nucleotide polymorphism rs134351, compared with homozygous carriers (n=15) of the major allele. (a–d) Significance testing of the difference between means was performed using the Mann–Whitney U test.
Increased IL-17C in the serum of UC patients may result from increased intestinal IL-17C due to colonic mucosal inflammation. Therefore we collected colonic specimen of macroscopically inflamed and not inflamed lesions of IBD patients (UC: n=23; CD: n=52). Quantification of IL-17C mRNA in these colonic lesions showed that IL-17C transcript is significantly increased in sites of inflammation (16.04 (4.00–28–08), arbitrary units) compared with non-inflamed sites (1.96 (0.85–3.07), arbitrary units) (P=0.005; Figure 1b). IL-17C mRNA was detected in 40.0% of all specimens, and was present in 51.5% of the inflamed vs. 28.3% of the not inflamed lesions. A significant increase of IL-17C mRNA in inflamed lesions was also seen in a subgroup analysis of CD patient samples only (6.1 fold increase vs. not inflamed, P<0.001; n=52) but did not reach statistical significance in the subgroup of UC patients (8.2 fold increase vs. not inflamed, P>0.05, n=23). The specific IL-17C receptor subunit IL-17RE mRNA levels in inflamed lesions did not significantly differ from those in not inflamed lesions (not shown). In summary, the differential IL-17C mRNA and serum protein levels suggest an involvement of IL-17C in the pathogenesis of intestinal inflammation during human IBD.
Modulation of IL-17C serum levels by IL23R genotype status and clinical characteristics of IBD patients
In the next step, we analyzed whether IL-17C serum levels correlate with clinical characteristics of the analyzed IBD patients (n=125). No statistically significant differences regarding IL-17C serum levels and disease activity (Crohn’s disease activity index or colitis activity index) and C-reactive protein serum levels were found (Supplementary Table S1 online). Given the well-established involvement of Th17 cells in the pathogenesis of IBD, especially CD, we genotyped the UC and CD patients; the results of which were used for IL-17C serum level measurements, for five IBD risk-increasing and five IBD risk-decreasing IL23R single-nucleotide polymorphisms,31, 32 which were previously reported to modulate Th17 cytokine levels.33, 34 No significant correlation was found between IL-17C serum levels in CD and UC patients and the odds ratios (OR) of the respective IL23R single-nucleotide polymorphisms (not shown). However, UC patients who carry one or two minor allele(s) of the IBD risk-decreasing IL23R single-nucleotide polymorphism rs134351 showed significantly lower IL-17C serum levels than homozygous major allele carriers (Figure 1c). This suggests that IL-23R signaling, which is crucial for promoting Th17-type inflammation, may also modulate IL-17C levels.
IL-17C mRNA levels significantly correlate with Th17 cytokine mRNA levels
The IL-23R-dependent modulation of IL-17C serum levels suggests a cross-regulation between Th17 cytokines and IL-17C. Similar to previous reports,35, 36, 37, 38, 39 we found increased levels of Th17-related cytokines and chemokines (IL-17A, IL-22, C-C motif chemokine ligand 20 (CCL20)) in inflamed lesions of our CD and UC patient cohort, as well as increased tumor necrosis factor (TNF)-α mRNA (Table 1). Given these increased cytokine levels, we analyzed to what extent IL-17C mRNA expression in IBD patients correlates with the mRNA expression levels of IL-17A, IL-22, IL-23, TNF-α, and CCL20. Significant correlations of IL-17C mRNA were found with IL-17A, TNF-α, CCL20, IL-23, and IL-22 mRNA (P<0.01) (Figure 2a–e). This demonstrates that IL-17C mRNA is increased simultaneously with Th17 cell–related inflammatory mediators and TNF-α mRNA in the inflamed intestine of IBD patients, indicating its participation in Th17 cell–mediated disease.
Interleukin (IL)-17C mRNA levels correlate with T helper type 17-related cytokine levels. (a–e) Correlation plots of arbitrary, absolute cytokine mRNA levels (calculated using the standard curve method and normalized to the median mRNA levels of HPRT1, YWHAZ, and RPL13A) in colonic biopsies of IBD patients. r and P-values were calculated using the Spearman correlation, determined by the program SPSS. Note that only biopsies with detectable IL-17C mRNA were used for the correlations (n=30). CCL20, C-C motif chemokine ligand 20; TNF, tumor necrosis factor.
IL-17C staining intensity is increased in colonic lesions of patients with active IBD
The experiments described above show increased IL-17C mRNA and protein levels in IBD patients. However, the precise cellular source of IL-17C in the inflamed human intestine is currently unknown. We therefore analyzed IL-17C protein levels in human colonic sections of IBD patients by immunohistochemistry.
For this, we validated a panel of commercially available anti-IL-17C polyclonal antibodies. Two of these antibodies (goat, R&D Systems and mouse, Sigma-Aldrich) did not stain the whole epithelium, but mostly single epithelial cells (Figure 3a,b) resembling the staining pattern of chromogranin A-positive enteroendocrine cells (Figure 3c). This pattern was also observed after staining with the third antibody (rabbit, Abcam), which in addition stained other more abundant epithelial cells (Figure 3d,e). In accordance with the pilot experiments demonstrating a strong production of IL-17C in HT-29 cells (which mainly consist of the goblet and absorptive enterocyte cell type), we decided to use the latter antibody for subsequent stainings. Important to note, IL-17C was present apically (facing the lumen) as well as basally (facing the lamina propria) in distinct crypt epithelial cells (Figure 3d,e), which was confirmed by immunofluorescence staining (Figure 3f,g). IL-17C-positive lamina propria cells were not found in the stained sections, and staining with the appropriate rabbit control immunoglobulin G fraction showed no staining at all (Supplementary Figure S1A).
Polarized interleukin (IL)-17C staining in intestinal epithelial cell of inflammatory bowel disease (IBD) patients. Representative immunohistochemical stainings (3,3′-diaminobenzidin-tetrahydrochloride (DAB) of IL-17C in colonic sections of IBD patients. (a, b) IL-17C staining using two different antibodies (from R&D Systems and Sigma-Aldrich, respectively). (c) Staining pattern of chromogranin A in the colon of IBD patients. Staining with anti-IL-17C antibody from Abcam demonstrated (d, f) basal (facing the lamina propria) and (e, g) apical (facing the crypt lumen) staining of distinct epithelial cells with DAB and AlexaFluor-546 staining. Magnification: × 40 (a–e); 2 × 63 (f); × 63 (g). Red arrows indicate positive staining.
Overall, semiquantitative analysis (CD active: n=9; CD remission: n=4; UC active: n=4; UC remission n=3; healthy controls: n=7) demonstrated significantly increased IL-17C staining intensity in combined IBD patients (CD+UC) (P=0.004), patients with active IBD (P=0.006), combined IBD patients in remission (P=0.015), CD patients with active disease (P=0.017), UC patients with active disease (P=0.007), and UC patients in remission (P=0.0003), compared with healthy controls (Figure 4a). Representative images of IBD patients are shown for CD patients with active disease (Figure 4b,c), CD patients in remission (Figure 4d,e), UC patients with active disease (Figure 4f,g), and UC patients in remission (Figure 4h,i), as well as representative images of healthy controls (Figure 4j,k). Frequently, weak staining of the colonic epithelial cells was also observed in healthy controls (Figure 4j).
Colonic interleukin (IL)-17C staining is increased in active inflammatory bowel disease (IBD). (a) Semiquantitative analysis of IL-17C in all stained sections of IBD patients and healthy controls: 0=no staining, 1=weak staining, 2=moderate staining and 3=strong staining (A=active disease; R=remission; Mean±s.e.m., *P<0.05 vs. healthy). (b–k) Representative immunohistochemical stainings of IL-17C in colonic sections of IBD patients and healthy controls (b, c: Crohn’s disease (CD) active disease; d, e: CD remission; f, g: ulcerative colitis (UC) active disease; h, i: UC remission; j, k: healthy control). Magnification: × 20.
Enteroendocrine chromogranin A-positive cells produce basal IL-17C, while goblet cells are major producers of apical IL-17C in the colon of IBD patients
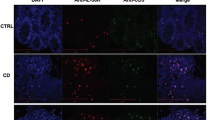
In order to identify the cell types that produce IL-17C in the inflamed colon of IBD patients, we performed double-immunofluorescence analysis of epithelial cells combined with confocal laser scanning microscopy (imaging of 1-μm optical slices). As mentioned above, the basal staining pattern of IL-17C in single epithelial cells resembled that of chromogranin A staining pattern. Accordingly, the simultaneous double-immunofluorescent detection of chromogranin A and IL-17C demonstrated overlapping chromogranin A and IL-17C staining, particularly in the basal region of single epithelial cells (Figure 5a).
Chromogranin A-positive enteroendocrine cells stain basally for interleukin (IL)-17C, whereas Mucin-2 (MUC2)-positive goblet cells stain apically for IL-17C in inflamed colonic lesions of inflammatory bowel disease patients. Representative double-immunofluorescence labeling (magnification: × 63) of IL-17C (AlexaFluor-546, red) with either (a) chromogranin A (CHGA) or (b) MUC2 (AlexaFluor-488, green) (left: single channels, middle: overlay, right: further magnification with confocal zoom ( × 7)). For each antigen (CHGA and MUC2), at least three colonic sections of each disease category were stained, demonstrating similar staining patterns. (c–j) Representative images of the different disease subgroups (c, g: Crohn’s disease (CD) active disease; d, h: CD remission; e, i: ulcerative colitis (UC) active disease; f, j: UC remission). (k, l) IL-17A and tumor necrosis factor (TNF)-α induce IL-17C expression in GOT1 and BON1 cells (quantitative real-time PCR). (m, n) IL-17C protein is only secreted by GOT1 cells and not by BON1 cells (enzyme-linked immunosorbent assay). a=significant vs. unstimulated control, b=significant vs. TNF-α stimulated.
IL-17C staining was not only observed basally but also apically in distinct epithelial cells, which morphologically resembled goblet cells. To verify IL-17C production by goblet cells, we co-stained IL-17C with the goblet cell marker Mucin-2 (MUC2). These stainings showed that apical IL-17C production occurs in MUC2-positive cells (Figure 5b). Again, not all MUC2-positive cells stained also positive for IL-17C, but apical IL-17C-positive staining was always positive for MUC2 staining; note that the anti-MUC2 antibody is directed against the precursor of MUC2, therefore the staining locates near the nucleus of the epithelial cell, rather than in the most apical granula, and for this reason does not overlap with IL-17C staining in the same cell (Figure 5b, right, magnified image). Representative images of IL-17C colocalization with chromogranin A and MUC2 are shown in Figure 5c–j for each analyzed IBD category (CD remission and active; UC remission and active).
Together, these findings demonstrate that basal IL-17C is produced by enteroendocrine cells, whereas apical IL-17C production occurs in vacuoles of goblet cells in the inflamed colon of IBD patients.
Autocrine action of IL-17C on IECs
IL-17C has been reported to act in an autocrine manner on epithelial cell lines, with most reports showing its action in human primary keratinocytes.14, 30 Recently, we also demonstrated upregulation of beta-defensin 2 in human primary keratinocytes by IL-17C stimulation.40 Reynolds et al.15 and Song et al. 41 demonstrated upregulation of tight junction components and apoptosis mediators in murine IECs by IL-17C. However, studies using human IECs are very limited, to date only demonstrating mitogen-activated protein (MAP) kinase activation in HT-29 cells upon IL-17C simulation.13 Therefore, we aimed to investigate further effects of IL-17C on human IECs using different cell lines.
First, we analyzed the expression of the IL-17C-specific receptor subunit, IL-17RE, on various IEC lines. IL-17RE mRNA and protein were present on the IEC lines HT-29, HCT116, SW480, DLD-1, and T84 (Supplementary Figure S1B,C). This was confirmed by quantitative real-time PCR (qRT-PCR) measurements, demonstrating high expression of IL-17RE mRNA in IECs, in contrast to isolated suspension and adherent peripheral blood monocyte cells (PBMCs), stimulated with the cytokines as indicated (Supplementary Figure S1D).
In a second step, we tested how IECs respond to IL-17C stimulation by functional assays. However, we could not detect MAP kinase (extracellular signal–regulated kinase (ERK)-1/2 and p38) phosphorylation in HT-29, HCT116, or DLD-1 cells by IL-17C stimulation (not shown). Furthermore, no effect on the cell proliferation of HT-29 cells (Supplementary Figure S1E) was detected, as well as no increased wound-healing capacity of HCT-116 cells after stimulation with IL-17C (Supplementary Figure S2A). Several reports suggested that IL-17C acts in concert with IL-22 and TNF-α on IECs.13, 14 Therefore, we stimulated HT-29 cells with IL-17C, IL-22, TNF-α, and the combination of these cytokines and measured their effect on the expression of several target genes (CXCL1, CXCL8, CCL20, DEFB4A, CLDN1, CLDN2, CLDN4, OCLN, ZO1, DEFB1, LCN2). Among these genes, we only detected a synergistic increase of CCL20 mRNA by stimulation with IL-17C in addition to TNF-α (Supplementary Figure S2B); however, this effect was only observed in HT-29 cells that were previously deprived of glucose, which promotes their differentiation into a absorptive enterocyte–like cell type,42 and in CaCo-2 cells (Supplementary Figure S2C). Furthermore, this effect was not routinely observed, probably due to the low magnitude (fold change=2). IL-17C stimulation was also tested on other IEC lines (HCT116, SW480, DLD-1, T84), without a detectable effect on the mentioned target gene expressions; these results were the same with recombinant human IL-17C from three different vendors (not shown).
In summary, only highly differentiated IEC lines were responsive to IL-17C stimulation and only in combination with other cytokines, such as TNF-α, resulting in an increased CCL20 expression which is the major chemoattractant for Th17 cells in the inflamed intestine.43
IL-17A amplifies the TNF-α-mediated secretion of IL-17C in enteroendocrine and goblet cells
To confirm the production of IL-17C in colonic enteroendocrine and goblet cells demonstrated in our immunohistochemical experiments (Figures 3, 4 and 5), we performed further experiments in appropriate intestinal cell lines. Given that both IL-17A and TNF-α are elevated in IBD39, 44 and their mRNA levels significantly correlate with IL-17C mRNA levels in our IBD patient samples (Figure 2), we hypothesized that IL-17A might synergize with TNF-α for the production of IL-17C in human IECs.
The GOT1 cell line represents a human neuroendocrine cell line originating from a human midgut carcinoid tumor,45 in contrast to, e.g., the pancreas-derived BON1 cell line. First, we used both the GOT1 and BON1 cell line for the stimulation experiments, in order to see whether IL-17C production is restricted to endocrine cells of the intestine (i.e., GOT1 cells) or also produced by endocrine cells of other tissues, such as the pancreas (i.e., BON1 cells). Combined stimulation with IL-17A+TNF-α (but not IL-17A or TNF-α alone) induced the expression of IL-17C mRNA in both GOT1 and BON1 cells (Figure 5k,l). However, only intestinal-derived GOT1 cells but not BON1 cells secreted IL-17C protein (Figure 5m,n). The HT-29 cell line is a well-established IEC model, consisting of goblet-like enterocytes expressing MUC2 under standard culture conditions.46 Similar to GOT1 cells, IL-17A+TNF-α induced a strong production of IL-17C in HT-29 cells, with IL-17A amplifying TNF-α-induced IL-17C mRNA (Figure 6a).
Interleukin (IL)-17A amplifies the tumor necrosis factor (TNF)-α-induced expression of IL-17C in intestinal epithelial cells. (a, b) quantitative real-time PCR measurements of IL-17C mRNA fold change after 6 h in HT-29 cells, normalized to glyceraldehyde 3-phosphate dehydrogenase mRNA levels. (c, d) Enzyme-linked immunosorbent assay (ELISA) measurement of IL-17C secretion after 48 h from HT-29 cells. IL-17R-neutralizing antibody was incubated 30 min before stimulation with cytokines. (e) Immunoblot detecting IL-17C protein in the cytoplasmic fraction of HT-29 cells (loading control: β-actin). (f) Kinetics of secreted IL-17C from HT-29 cells, measured by ELISA. Mean±s.e.m. are shown (n=3), and experiments were repeated at least three times. a=significant vs. unstimulated control, b=significant vs. TNF-α stimulated, c=significant vs. IL-17A+TNF-α stimulated (without antibody) (Student’s t-test P<0.05). (g) Immunofluorescent staining of cytoplasmic IL-17C in HT-29 cells, stimulated with the indicated cytokines for 4 h.
Because GOT1 cells grow relatively slowly with a long doubling time (>18 days), we used HT-29 cells (as they contain goblet-like cell clones46) for further experiments requiring high cell numbers. We confirmed specific IL-17A-mediated amplification of IL-17C in HT-29 cells by antibody-mediated neutralization of the necessary receptor subunit of the IL-17A receptor, IL-17RA. This resulted in decreased IL-17C transcript levels after stimulation with IL-17A and TNF-α, compared with the stimulation without antibody inhibition (Figure 6b). This effect was confirmed on protein level after 48 h of stimulation (Figure 6c,d). Western blotting experiments of the cytoplasmic fraction of HT-29 cells indicated that IL-17C protein accumulated in the cytoplasm until 2–4 h and then slightly decreased again (Figure 6e). This was in accordance with time course experiments showing that IL-17A further amplifies secreted IL-17C protein as soon as 8 h (Figure 6f) compared with TNF-α stimulation alone. Immunofluorescent staining of IL-17C in HT-29 cells confirmed the increased cytoplasmic IL-17C production in HT-29 cells after IL-17A and TNF-α stimulation (Figure 6g). The increased expression of IL-17C and CEBP transcription factors upon combined IL-17A and TNF-α stimulation was exemplarily confirmed in a subclone of HT-29 cells, which consists mainly of goblet-like cells (HT-29 MTX, Supplementary Figure S2D–F).
Together, these experiments demonstrate that enteroendocrine cells (GOT1) and goblet cells (HT-29) produce IL-17C, confirming our immunohistochemical data. IL-17A strongly amplifies IL-17C secretion from IECs in a TNF-α-dependent manner. Importantly, only endocrine cells of intestinal origin (GOT1), and not endocrine cells of pancreatic origin (BON1), produced IL-17C upon cytokine stimulation, suggesting a unique role of IL-17C restricted to the inflamed intestine of IBD patients.
IL-17C secretion from IECs by combined IL-17A and TNF-α stimulation is abrogated by the inhibition of ACT1, nuclear factor (NF)-κB, MAP kinases, and Akt and counter-regulated by the RNase monocyte chemotactic protein–induced protein 1 (MCPIP)
Currently, there are no reports investigating the molecular pathways leading to IL-17C release from human IECs. Therefore, we aimed to identify signaling mediators that are crucial for IL-17A- and/or TNF-α-induced IL-17C production in HT-29 cells. ACT1 is a necessary adaptor protein for IL-17A-induced expression of inflammation-related gene targets.47 Consequently, siRNA-mediated knockdown of ACT1 reversed the amplification of IL-17C mRNA expression by IL-17A (Figure 7a). Both IL-17A and TNF-α are known to activate MAP kinases and Akt, as well as NF-κB.48 We examined the effect of NF-κB pathway inhibition on the IL-17A+TNF-α-induced IL-17C mRNA expression from HT-29 cells. Pretreatment with the IKK-2 inhibitor SC-514 strongly impaired IL-17C mRNA expression after 6 h of IL-17A and TNF-α stimulation (Figure 7b). Further experiments with pharmacological kinase inhibitors revealed that ERK-1/2, p38, and Akt are also fundamentally involved in the secretion of IL-17C from IECs upon stimulation with IL-17A and TNF-α (Figure 7c). Importantly, IL-17A alone did neither activate NF-κB in IECs nor significantly increase the NF-κB activation by TNF-α in electrophoretic mobility shift assay experiments (Supplementary Figure S3A); furthermore, immunoblot experiments showed that IL-17A did not increase TNF-α-induced phosphorylation of ERK-1/2 and p38 kinases (not shown). Based on this, we hypothesize that the repressive effect of PD98059, SB203580, and SC-514 on the induced expression of IL-17C likely arises from the impairment of TNF-α-induced MAP kinase and NF-κB activation and not via IL-17A-activated pathways.
Combined interleukin (IL)-17A+TNF (tumor necrosis factor)-α-induced IL-17C production in intestinal epithelial cells is dependent on ACT1, nuclear factor-κB, mitogen-activated protein kinase, Akt, and monocyte chemotactic protein–induced protein 1 (MCPIP). (a, b) IL17C mRNA fold change in HT-29 cells after 6 h of stimulation, preincubated with either non-targeting control siRNA or ACT1 siRNA for 24 h or SC-514 for 30 min and normalized to glyceraldehyde 3-phosphate dehydrogenase mRNA levels (c) IL-17C secretion from HT-29 cells after 48 h measured by enzyme-linked immunosorbent assay after pretreatment with the indicated kinase inhibitors for 30 min (50 μM each). (d) Western blotting of phosphorylated Akt in HT-29 cells after 30 min of stimulation (total Akt served as a loading control). (e) Zc3h12a (MCPIP) mRNA fold change in HT-29 cells after 6 h. (f) IL-17C secretion from HT-29 cells after 24 h, pretreated with control or Zc3h12a (MCPIP) siRNA. All experiments (mean±s.e.m., n=3) were repeated at least three times. a=significant vs. IL-17A+TNF-α stimulated (control siRNA or no inhibitor, respectively), b=significant vs. TNF-α stimulated (no inhibitor), c=significant vs. unstimulated control (Student’s t-test, P<0.05). Successful siRNA-mediated knockdown of ACT1 and Zc3h12a was confirmed by quantitative real-time PCR.
Therefore we analyzed further mediators that might be involved in the specific transduction of IL-17A-induced signals. IL-17A further increased the phosphorylation of Akt compared with sole TNF-α-stimulation (Figure 7d). In line with this, blockade of the Akt kinase pathway resulted in the strongest inhibition of combined IL-17A- and TNF-α-mediated IL-17C secretion (Figure 7c). Another protein that is synergistically increased by IL-17A and TNF-α in IECs is MCPIP (Zc3h12a; Figure 7e). MCPIP has been implicated in the sequestration of the cytokine IL-6 in monocytes.49 Therefore we analyzed whether MCPIP might be also involved in the sequestration of cytokines in IECs, i.e. IL-17C. Interestingly, siRNA-mediated knockdown of MCPIP mRNA resulted in a strong increase (20-fold compared with control siRNA) of IL-17A- and TNF-α-induced IL-17C secretion from IECs (Figure 7f), suggesting a repressive mechanism on IL-17C production involving MCPIP, which is triggered only by combined IL-17A and TNF-α stimulation of IECs and not by TNF-α stimulation alone. Taken together, these results indicate that ACT1, MAP kinases, Akt, NF-κB, and MCPIP are crucial regulatory components of IL-17A- and TNF-α-induced IL-17C production in IECs, with ACT1, Akt, and MCPIP representing signaling mediators specifically and/or additionally activated by IL-17A stimulation.
IL-17A-mediated amplification of IL-17C release from IECs is dependent on CCAAT/enhancer-binding protein family members but not mRNA stabilization
IL-17A is known to stabilize the mRNA half-life time of cytokines and chemokines, e.g. CXCL1 (C-X-C motif chemokine ligand 1).50 Therefore we hypothesized that the stimulatory effect of IL-17A on IL-17C expression might be a result of transcript stabilization, i.e. prolonged half-life of the IL-17C mRNA. Our experiments with the mRNA synthesis inhibitor actinomycin D (Act D) showed that IL-17A prolonged the half-life of CXCL1 mRNA (Figure 8a). However, IL-17A did not significantly influence IL-17C mRNA stability, compared with stimulation with TNF-α alone (Figure 8b). We further analyzed the involvement of transcription factors that are synergistically induced by IL-17A and TNF-α. In silico transcription factor binding site search (TFSEARCH, http://www.cbrc.jp/research/db/TFSEARCH.html) in the promoter region of IL17C predicted binding sites for the transcription factors C/EBP-α (+62 to +74) and C/EBPβ (−1507 to −495) (threshold score for binding site prediction: 85.0). Although C/EBPα mRNA levels did not significantly change after 6 h of combined IL-17A and TNF-α stimulation (Supplementary Figure S3B), the transcription factors C/EBPβ and C/EBPδ of the same protein family were synergistically induced (Supplementary Figure S3C,D), with the highest fold change seen for C/EBPδ mRNA (Figure 8c). Accordingly, we detected nuclear accumulation of C/EBPδ in HT-29 cells as soon as 2 h of stimulation with IL-17A and TNF-α (Figure 8d). C/EBPα, C/EBPβ, and C/EBPδ are known to act in homo- and hetero-dimers and bind the same DNA sequence (e.g. Kinoshita et al.51). Therefore, we hypothesized that C/EBPδ might regulate IL-17C expression too, although binding sites in the IL-17C promoter were only predicted for C/EBPα and C/EBPβ.
Interleukin (IL)-17A-mediated amplification of IL-17C release from intestinal epithelial cells is dependent on CCAAT/enhancer-binding protein family members but not mRNA stabilization. (a, b) C-X-C motif chemokine ligand 1 (CXCL1) and IL-17C mRNA decay in HT-29 cells after the treatment with 10 μM actinomycin D. (c) CEBPD mRNA fold change in HT-29 cells after 6 h (a=P<0.05 vs. unstimulated and tumor necrosis factor (TNF)-α stimulated, Student’s t-test). (d) Western blotting of C/EBPδ in the nuclear fraction of HT-29 cells (p85N5 served as the loading control). (e) Intracellular IL-17C protein and (f) secreted IL-17C is increased in IL-17A+TNF-α-stimulated HT-29 cells when C/EBPβ and C/EBPδ are overexpressed. (g) IL-17C secretion after 48 h from HT-29 cells, pretreated with the siRNAs for 24 h as indicated. Mean±s.e.m. (n=3) is shown; experiments were repeated three times. *significant vs. mock transfection with the respective empty control vector (“empty vector β” and “empty vector δ”). (h, i) siRNA-mediated knockdown (24 h) of C/EBPβ and C/EBPδ and subsequent stimulation with IL-17A+TNF-α for 6 h upregulates the expression of the respective other C/EBP isoform. a=significant vs. IL-17A+TNF-α stimulated (control siRNA), b=significant vs. unstimulated control (control siRNA), c=significant vs. TNF-α stimulated (control siRNA) (Student’s t-test, P<0.05).
Given the IL-17A-mediated induction of C/EBPβ and C/EBPδ, we tested in a further step whether overexpression and knockdown of C/EBPs affects IL-17C production by IECs. Therefore, we transfected HT-29 cells with C/EBPβ or C/EBPδ overexpression plasmids or the according empty control vectors. Successful overexpression of C/EBPβ or C/EBPδ was verified using qRT-PCR (Supplementary Figure S3E,F). In IL-17A+TNF-α-stimulated HT-29 cells, both C/EBPβ and C/EBPδ overexpression significantly increased intracellular IL-17C (Figure 8e) and secreted IL-17C (Figure 8f), compared with the cells transfected with the respective empty control plasmids (“empty vector β” and “empty vector δ”, respectively). Overexpression of C/EBPβ or C/EBPδ did not affect IL-17C protein levels, indicating that additional mediators stimulated by IL-17A and TNF-α are necessary for the production of IL-17C protein in HT-29 cells.
In accordance with the overexpression experiments, simultaneous siRNA-mediated knockdown of both C/EBPβ and C/EBPδ resulted in decreased IL-17C secretion from HT-29 cells (Figure 8g). We used a simultaneous knockdown of both C/EBPβ and C/EBPδ, because previous experiments had shown that siRNA-mediated knockdown of either C/EBPβ or C/EBPδ increased the mRNA expression of the respective other C/EBP family member upon combined IL-17A and TNF-α stimulation (Figure 8h,i), suggesting functional redundancy of C/EBPβ and C/EBPδ. This functional redundancy is consistent with previous reports.52
In summary, these experiments show that the synergistic IL-17C production by IL-17A and TNF-α in human IECs is dependent on C/EBPβ- and C/EBPδ-regulated transcription and not mRNA stabilization, as previously observed for other IL-17A-induced cytokines and chemokines.
DISCUSSION
IL-17C is a novel member of the IL-17 cytokine family. However, its role in human intestinal inflammation is currently unknown, given that previous experiments focused exclusively on murine models of inflammation. Moreover, the detailed signal transduction leading to increased IL-17C in human IECs is unknown. In mice, the source of IL-17C has been identified to be epithelial cells, but the exact cellular origin and its sub-cellular regulation in the inflamed intestine is unknown. This study represents the first report of the role of IL-17C in human IBD and its association with other Th17 cytokines that contribute to the disease pathology in human IBD.
We demonstrated the pathological involvement of the IL-17C pathway in human IBD by (1) elevated IL-17C serum levels in IBD patients; (2) increased IL-17C mRNA in inflamed colonic lesions of IBD patients; and (3) increased IL-17C staining in colonic sections of active IBD, compared with healthy controls and patients in remission. Increased colonic IL-17C mRNA in inflamed lesions of UC patients is supported by Im et al.,17 who showed that IL-17C mRNA is increased in UC compared with healthy controls.
Strikingly, our immunohistochemical experiments showed that enteroendocrine cells, along with goblet cells, are the major producers of IL-17C in the inflamed intestine of IBD patients. IL-17C production by chromogranin A-positive cells may provide a potential link between the increased IL-17C levels in IBD patients and reported higher serum levels of chromogranin A in IBD patients, compared with healthy controls.53 Moreover, given that a major source of intestinal IL-17C production in IBD are enteroendocrine cells, our study provides novel insights of how the neuroendocrine system may interact with the Th17 pathway and thereby influence intestinal inflammation during IBD (reviewed in Bonaz and Bernstein54). This is of great interest, given that several studies reported a role of psychosocial stress mediated partially by the neuroendocrine system as major contributor for IBD onset and IBD flares (reviewed in Bonaz and Bernstein54).
The interaction of the neuroendocrine and Th17 pathways is further emphasized by our data showing a significant correlation of IL-17C with Th17-related inflammatory mediators, which are well-established mediators of the disease pathogenesis of human IBD. The ability of IL-17A to increase the TNF-α-induced production of IL-17C in enteroendocrine and goblet cells, as demonstrated here, strongly suggests its involvement in a Th17-mediated disease course in IBD.
Regarding the effector functions of IL-17C, we detected its receptor expression (IL-17RE) on IECs and an increased expression of CCL20 mRNA upon stimulation with IL-17C in addition to TNF-α. This supports the participation of IL-17C in a Th17-dominant IBD disease course, as CCL20 is a potent chemoattractant for Th17 cells.43 Moreover, we previously demonstrated increased CCL20 expression in active IBD.38 However, no effect on MAP kinase activation and the analyzed target gene expressions was detected in various transformed IEC lines (similar results were reported by Rajita Pappu, Department of Immunology, Genentech, San Francisco, CA, personal communication) and with recombinant human IL-17C from three different vendors. Furthermore, there was no effect on the proliferation and wound-healing capacity of IECs. Therefore we hypothesize that the major IL-17C-responsive target cells might be other cells than epithelial cells in the human intestine. Importantly, Chang et al.16 reported that IL-17RE is selectively expressed on Th17 cells and that IL-17C potentiates Th17 responses. Thus, epithelial-produced IL-17C may represent an important mediator of epithelial–immune cell (Th17) cross-talk. Another possibility for the lacking responsiveness to IL-17C stimulation is that we used transformed and not primary IEC lines; however, e.g., IL-17A stimulation of the same cell lines yields consistent and dose-dependent results (see experiments above and Friedrich et al.40). Finally, the autocrine effects of IL-17C seen in murine IECs, in contrast to weak effects in human IECs, may be attributed to huge differences between murine and human physiology and/or differences in the potency of different IL-17 family members. For example, IL-17A demonstrates a 30-fold stronger biological effect than IL-17F of the same cytokine family (reviewed in Gaffen et al.55).
Finally, no reports exist about the pathways that regulate IL-17C production in IECs. We uncovered molecular mediators (NF-κB, ERK-1/2, p38, Akt, MCPIP, C/EBPβ, and C/EBPδ) that are decisively involved in the regulation of IL-17C release from IECs upon combined stimulation with IL-17A and TNF-α. Our experiments suggest ACT1, Akt, MCPIP, and C/EBP family members as specific mediators of IL-17A signaling that are profoundly involved in the regulation of IL-17C production in IECs. These mediators may represent valuable targets for the intervention in IL-17A-dependent modification of intestinal IL-17C levels in colonic inflammation.
In summary, our findings support the involvement of IL-17A-amplified epithelial IL-17C production in active human IBD and reveal a novel inter-regulation of the neuroendocrine system with the Th17 pathway.
METHODS
Cytokines. Recombinant human (rh) IL-17A, IL-17C, IL-22, TNF-α, transforming growth factor-β, granulocyte-macrophages colony-stimulating factor, and IL-4 were purchased from R&D Systems (Minneapolis, MN), IL-12, IL-23, and IL-6 from eBioscience (Frankfurt, Germany). rhIL-17C was additionally obtained from eBioscience and Abnova (Heidelberg, Germany). If not stated otherwise, IL-17A, IL-17C, and IL-22 were used at a concentration of 100 ng ml−1. TNF-α was used at 50 ng ml−1.
Cell culture. The human colorectal cancer-derived IEC lines HT-29, HCT116, SW480, DLD-1, and T84 (obtained from American Type Culture Collection, Rockville, MD) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (Sigma-Aldrich, Munich, Germany) at 37 °C in a humidified 5% CO2 atmosphere, BON1 cells were cultured in Dulbecco’s modified Eagle’s medium/F12 (1:1) medium (10% fetal calf serum and 1% penicillin/streptomycin), and the GOT1 cell line45 (kindly provided by Professor Ola Nilsson, Sahlgrenska University Hospital, Gothenburg, Sweden) was maintained in RPMI medium, including 10% fetal calf serum, 1% penicillin/streptomycin, 0.135 IU ml−1 insulin, and 5 mg dl−1 apotransferrin. For all experiments, cells were allowed to attach overnight and then treated as indicated.
Biopsy isolation and serum collection. Human colonic tissue was obtained from patients undergoing colonoscopy. The study was approved by the Ethics committee of the Ludwig-Maximilians-University Munich (Department of Medicine) and adhered to the ethical principles for medical research involving human subjects of the Helsinki Declaration. Prior to intestinal biopsy sampling, all participating patients gave written, informed consent. Biopsies were routinely taken with a depth not exceeding the mucosal layer of the intestine and immediately stored in RNAlater (Qiagen, Hilden, Germany). Human blood and serum samples were obtained after written informed consent from patients and controls, aliquoted and stored at −80 °C until further analysis.
PBMC isolation. PBMCs were isolated using Vacutainer collection tubes (Becton Dickinson, Heidelberg, Germany) following the manufacturer’s instructions. Briefly, adherent and suspension PBMC fractions were separated by plastic adherence in serum-free medium. Then adherent PBMCs were stimulated with the cytokines as indicated for 7 days, with medium exchange every 3 days. Suspension PBMCs were cultured in RPMI supplemented with anti-CD3 and anti-CD28 (eBioscience), and the cytokines as indicated, for 5 days.
Protein isolation, gel electrophoresis, and immunoblot. Cells were solubilized in a buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% Nonidet P-40, 2 mM phenylmethylsulfonyl fluoride, protease, and phosphatase inhibitor cocktail (Roche, Mannheim, Germany) and cleared by centrifugation at 18,000 g. Total protein was separated on a polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Thermo Fisher Scientific, Bonn, Germany). After membrane blocking in 3% bovine serum albumin, primary antibodies (anti-phospho-Akt 1:1000 and β-actin 1:1000 (Cell Signaling, Danvers, MA); 1 μg ml−1 anti-IL-17C (R&D Systems); 0.4 μg ml−1 anti-IL-17RE (Sigma-Aldrich; 1 μg ml−1) anti-C/EBPδ (Santa Cruz Biotechnology, Santa Cruz, CA); 1 μg ml−1 anti-p84N5 (Abcam, Cambridge, UK)) were incubated overnight at 4 °C, appropriate horseradish peroxidase–conjugated secondary antibodies (1:20.000, GE Healthcare, Buckinghamshire, UK) were incubated for 2 h at room temperature, and signals were detected using Pico Super Signal chemiluminescent substrate (Thermo Fisher Scientific, Bonn, Germany).
Nuclear extracts and electrophoretic mobility shift assay. Nuclear lysates were prepared by pelleting nuclei after cell lysis in buffer A (10 mM Hepes pH 7,9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.5% NP-40). Isolated nuclei were incubated 30 min on ice in buffer B (10 mM Hepes pH 7,9, 0.4 mM NaCl, 1 mM EDTA, 1 mM EGTA, 25% glycerol), and 5 μg of the nuclear extract (supernatant) were incubated with 50 fmol of 3′-biotinylated, double-stranded probes in the presence of 1 μg poly d(I)-d(C) for 30 min on ice. Samples were separated at 100 V on a 6% polyacrylamide gel and transferred (30 V, 1 h) in 0.5 × Tris-borate-EDTA buffer. Signal detection was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Bonn, Germany). The sequence for the NF-κB consensus probe was: 5′-AGTTGAGGGGACTTTCCCAGGC-3′.
Reverse transcriptase PCR and qRT-PCR. RNA was isolated using the RNeasy Mini Kit (Qiagen) and reverse transcribed (0.3 μg RNA) with the Transcriptor First Strand cDNA Synthesis Kit (Roche) using oligo-dT primer. PCR (40 cycles of denaturing at 95 °C for 30 s, annealing at 60–68 °C for 30 s, extension at 72 °C for 30 s) was run on a Roche LightCycler 480 using LightCycler 480 SYBR Green Master I (Roche). If possible, all primer pairs (purchased from TIB MOLBIOL, Berlin, Germany) were designed (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) to span intronic sequences in order to avoid amplification of genomic DNA. Primer sequences are listed in Table 2. If not stated otherwise, mRNA fold changes were manually calculated using ΔΔCT method and normalized to glyceraldehyde 3-phosphate dehydrogenase expression, and P-values were calculated using the unpaired, two-tailed Student’s t-test (n=3).
qRT-PCR of colonic biopsy samples. RNA from colonic biopsies was isolated using ceramic bead-assisted (Roche Diagnostics) homogenization, followed by RNA isolation using the RNeasy Mini Kit (Qiagen). Absolute mRNA values were calculated using a standard curve that was previously set up by serial dilutions of the isolated, pooled RNA. Samples were normalized to geometric means of the respective YWHAZ, HPRT1, and RPL13A mRNA levels.
Antibody neutralization, pharmacological inhibitor experiments, and assessment of mRNA stability concentrations. Cells were incubated with 5 μg ml−1 anti-IL-17R antibody (R&D Systems), the IKK-2 inhibitor SC-514 (25 μg ml−1), the MEK-1 inhibitor PD98059, the p38 inhibitor SB203580, or the phosphoinositide-3-kinase (PI3 kinase) inhibitor wortmannin (all from Tocris Bioscience, Bristol, UK, 50 μM each). Then cells were stimulated with the indicated cytokines, and RNA or protein was isolated as described above. For the determination of mRNA stability, cells were stimulated as indicated, and mRNA synthesis was stopped by the addition of 10 μM actinomycin D (Santa Cruz Biotechnology), followed by RNA isolation at the indicated time points.
Plasmid transfection and protein overexpression. C/EBPβ and C/EBPδ overexpression (CMV promoter) and empty control plasmids were purchased from Origene (Rockville, MD) and immediately used for transfection in HT-29 cells. Plasmid transfection was performed using Lipofectamine LTX reagent (Life Technologies, Darmstadt, Germany) according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were stimulated with cytokines, and RNA or protein was isolated as indicated. mRNA overexpression was confirmed by qRT-PCR.
RNA silencing. HT-29 cells were transfected with 10 nM non-targeting or Silencer Select siRNAs (Life Technologies) as indicated using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s instructions. The following day, cells were stimulated with cytokines as indicated. Efficient knockdown was assessed by qRT-PCR.
Immunohistochemistry. Paraffin-embedded sections were deparaffinized and subjected to heat-induced epitope retrieval (pH 9). After that, endogenous peroxidase was blocked (1% H2O2), followed by normal serum (5% normal serum) and avidin/biotin blocking. Primary antibodies (three different anti-IL-17C antibodies from Abcam, Sigma-Aldrich, and R&D Systems; anti-chromogranin A from Dako, Glostrup, Denmark) were incubated overnight at 4 °C. The next day, biotinylated secondary antibodies (Dako) were incubated at room temperature for 2 h, followed by 45-min incubation with avidin–biotin complex and 3,3′-diaminobenzidin-tetrahydrochloride substrate (both Vector Laboratories, Burlingame, CA) and counterstaining with Mayer’s haemalaun (Carl Roth, Karlsruhe, Germany). Images were acquired on a Zeiss Axioskop 40 microscope (Carl Zeiss Microscopy, Göttingen, Germany).
For double-immunoflourescence labeling, mixtures of primary antibodies (0.4 μg ml−1 anti-IL-17C from Abcam; 0.2 μg ml−1 anti-chromogranin A and 0,02 μg ml−1 anti-MUC2 from Dako) were applied, after heat-induced epitope retrieval (pH=9) and blocking in 3% bovine serum albumin, overnight at 4 °C. The next day, sections were incubated with fluorescence-labeled secondary antibodies (AlexaFluor488 and AlexaFluor546, Life Technologies; 4 μg ml−1), together with TO-PRO-3 (Life Technologies) as a nuclear counter-stain. Slides were mounted in ProLong Gold antifade (Life Technologies) and imaged on a confocal laser scanning microscope (LSM 510, Carl Zeiss), acquiring optical slices with a thickness of 1 μm if not stated otherwise.
Cell proliferation assay. Cell proliferation was assessed by water-soluble tetrazolium assay (Roche) as indicated by the manufacturer.
Enzyme-linked immunosorbent assay. Serum or cell culture supernatant was directly measured by ELISA or stored aliquoted at −80 °C, using the human IL-17C Duo Set ELISA (R&D Systems) according to the manufacturer’s instructions. Alternatively, the IL-17C ELISA Kit of Assay Biotech was used.
IL23R genotyping. Genomic peripheral blood leukocyte DNA was isolated using the DNA Blood Minikit from Qiagen. Ten IL23R single-nucleotide polymorphisms (rs10048019, rs7517847, rs10489629, rs2201841, rs11465804, rs11209026, rs1343151, rs10889677, rs11209032, rs1495965) were genotyped by PCR and melting curve analysis using a pair of FRET probes as previously described.8, 33, 56, 57
Statistics and data presentation. For in vitro experiments in HT-29 cells, P-values were calculated using two-tailed unpaired Student’s t-test. For all other experiments, the statistical test used is indicated in the corresponding figure legend. qRT-PCR fold change (ΔΔ cp method, relative fold change) or absolute value calculations (standard curve method, arbitrary values), correlations and statistics were either performed manually using the Microsoft Excel datasheets (Microsoft Corporation, Redmond, WA) or by using the programs qbase+ (version 2.5, http://www.biogazelle.com/qbaseplus) and SPSS (version 17.0, SPSS, Chicago, IL), as indicated. If not stated otherwise, data are given in mean values and round brackets indicate the 95% confidence interval.
References
Brand, S. Crohn's disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn's disease. Gut 58, 1152–1167 (2009).
Monteleone, I., Sarra, M., Pallone, F. & Monteleone, G. Th17-related cytokines in inflammatory bowel diseases: friends or foes? Curr. Mol. Med. 12, 592–597 (2012).
Abraham, C. & Cho, J.H. IL-23 and autoimmunity: new insights into the pathogenesis of inflammatory bowel disease. Annu. Rev. Med. 60, 97–110 (2009).
Abraham, C. & Cho, J. Interleukin-23/Th17 pathways and inflammatory bowel disease. Inflamm. Bowel Dis. 15, 1090–1100 (2009).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Anderson, C.A. et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 43, 246–252 (2011).
Glas, J. et al. Novel genetic risk markers for ulcerative colitis in the IL2/IL21 region are in epistasis with IL23R and suggest a common genetic background for ulcerative colitis and celiac disease. Am. J. Gastroenterol. 104, 1737–1744 (2009).
Michel, M.L. et al. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J. Exp. Med. 204, 995–1001 (2007).
Shibata, K., Yamada, H., Hara, H., Kishihara, K. & Yoshikai, Y. Resident Vdelta1+ gammadelta T cells control early infiltration of neutrophils after Escherichia coli infection via IL-17 production. J. Immunol. 178, 4466–4472 (2007).
Miossec, P. & Kolls, J.K. Targeting IL-17 and TH17 cells in chronic inflammation. Nat. Rev. Drug Discov. 11, 763–776 (2012).
Gaffen, S.L. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 9, 556–567 (2009).
Song, X. et al. IL-17RE is the functional receptor for IL-17C and mediates mucosal immunity to infection with intestinal pathogens. Nat. Immunol. 12, 1151–1158 (2011).
Ramirez-Carrozzi, V. et al. IL-17C regulates the innate immune function of epithelial cells in an autocrine manner. Nat. Immunol. 12, 1159–1166 (2011).
Reynolds, J.M., Martinez, G.J., Nallaparaju, K.C., Chang, S.H., Wang, Y.H. & Dong, C. Cutting edge: regulation of intestinal inflammation and barrier function by IL-17C. J. Immunol. 189, 4226–4230 (2012).
Chang, S.H., Reynolds, J.M., Pappu, B.P., Chen, G., Martinez, G.J. & Dong, C. Interleukin-17C promotes Th17 cell responses and autoimmune disease via interleukin-17 receptor E. Immunity 35, 611–621 (2011).
Im, E., Jung, J. & Rhee, S.H. Toll-like receptor 5 engagement induces interleukin-17C expression in intestinal epithelial cells. J. Interferon Cytokine Res. 32, 583–591 (2012).
Friedrich, M., Tillack, C., Wollenberg, A., Schauber, J. & Brand, S. IL-36gamma sustains a pro-inflammatory self-amplifying loop with IL-17C in anti-TNF-induced psoriasiform skin lesions of patients with Crohn’s disease. Inflamm. Bowel Dis. 20, 1891–1901 (2014).
Cario, E. & Podolsky, D.K. Differential alteration in intestinal epithelial cell expression of Toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect. Immun. 68, 7010–7017 (2000).
Ey, B. et al. Loss of TLR2 worsens spontaneous colitis in MDR1A deficiency through commensally induced pyroptosis. J. Immunol. 190, 5676–5688 (2013).
Hueber, W. et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700 (2012).
Hueber, W. et al. Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci. Transl. Med. 2, 52ra72 (2010).
Laroui, H. et al. Dextran sodium sulfate (DSS) induces colitis in mice by forming nano-lipocomplexes with medium-chain-length fatty acids in the colon. PLoS One 7, e32084 (2012).
Scheiffele, F. & Fuss, I.J. Induction of TNBS colitis in mice. Curr. Protoc. Immunol. Chapter 15 (Unit 15), 19 (2002).
Nenci, A. et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 446, 557–561 (2007).
Sturlan, S. et al. Interleukin-10-deficient mice and inflammatory bowel disease associated cancer development. Carcinogenesis 22, 665–671 (2001).
Powrie, F., Coffman, R.L. & Correa-Oliveira, R. Transfer of CD4+ T cells to C.B-17 SCID mice: a model to study Th1 and Th2 cell differentiation and regulation in vivo. Res. Immunol. 145, 347–353 (1994).
Seok, J. et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 110, 3507–3512 (2013).
Mestas, J. & Hughes, C.C. Of mice and not men: differences between mouse and human immunology. J. Immunol. 172, 2731–2738 (2004).
Johnston, A. et al. Keratinocyte overexpression of IL-17C promotes psoriasiform skin inflammation. J. Immunol. 190, 2252–2262 (2013).
Duerr, R.H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Glas, J. et al. rs1004819 is the main disease-associated IL23R variant in German Crohn's disease patients: combined analysis of IL23R, CARD15, and OCTN1/2 variants. PLoS One 2, e819 (2007).
Schmechel, S. et al. Linking genetic susceptibility to Crohn's disease with Th17 cell function: IL-22 serum levels are increased in Crohn's disease and correlate with disease activity and IL23R genotype status. Inflamm. Bowel Dis. 14, 204–212 (2008).
Diegelmann, J. et al. Intestinal DMBT1 expression is modulated by Crohn’s disease-associated IL23R variants and by a DMBT1 variant which influences binding of the transcription factors CREB1 and ATF-2. PLoS One 8, e77773 (2013).
Dambacher, J. et al. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut 58, 1207–1217 (2009).
Brand, S. et al. IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G827–G838 (2006).
Seiderer, J. et al. Role of the novel Th17 cytokine IL-17F in inflammatory bowel disease (IBD): upregulated colonic IL-17F expression in active Crohn's disease and analysis of the IL17F p.His161Arg polymorphism in IBD. Inflamm. Bowel Dis. 14, 437–445 (2008).
Brand, S. et al. Cell differentiation dependent expressed CCR6 mediates ERK-1/2, SAPK/JNK, and Akt signaling resulting in proliferation and migration of colorectal cancer cells. J. Cell Biochem. 97, 709–723 (2006).
Fujino, S. et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut 52, 65–70 (2003).
Friedrich, M., Diegelmann, J., Beigel, F. & Brand, S. IL-17A alone weakly affects the transcriptome of intestinal epithelial cells but strongly modulates the TNF-alpha-induced expression of inflammatory mediators and inflammatory bowel disease susceptibility genes. Inflamm. Bowel Dis. 20, 1502–1515 (2014).
Song, X. et al. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity 40, 140–152 (2014).
Zweibaum, A. et al. Enterocytic differentiation of a subpopulation of the human colon tumor cell line HT-29 selected for growth in sugar-free medium and its inhibition by glucose. J. Cell Physiol. 122, 21–29 (1985).
Hirota, K. et al. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J. Exp. Med. 204, 2803–2812 (2007).
Komatsu, M. et al. Tumor necrosis factor-alpha in serum of patients with inflammatory bowel disease as measured by a highly sensitive immuno-PCR. Clin. Chem. 47, 1297–1301 (2001).
Kolby, L. et al. A transplantable human carcinoid as model for somatostatin receptor-mediated and amine transporter-mediated radionuclide uptake. Am. J. Pathol. 158, 745–755 (2001).
Lesuffleur, T. et al. Differential expression of the human mucin genes MUC1 to MUC5 in relation to growth and differentiation of different mucus-secreting HT-29 cell subpopulations. J. Cell Sci. 106 (Pt 3), 771–783 (1993).
Qian, Y. et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 8, 247–256 (2007).
Gaffen, S.L. An overview of IL-17 function and signaling. Cytokine 43, 402–407 (2008).
Matsushita, K. et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190 (2009).
Sun, D., Novotny, M., Bulek, K., Liu, C., Li, X. & Hamilton, T. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). Nat. Immunol. 12, 853–860 (2011).
Kinoshita, S., Akira, S. & Kishimoto, T. A member of the C/EBP family, NF-IL6 beta, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc. Natl. Acad. Sci. USA 89, 1473–1476 (1992).
Ruddy, M.J. et al. Functional cooperation between interleukin-17 and tumor necrosis factor-alpha is mediated by CCAAT/enhancer-binding protein family members. J. Biol. Chem. 279, 2559–2567 (2004).
Sciola, V. et al. Plasma chromogranin a in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 15, 867–871 (2009).
Bonaz, B.L. & Bernstein, C.N. Brain-gut interactions in inflammatory bowel disease. Gastroenterology 144, 36–49 (2013).
Gaffen, S.L., Kramer, J.M., Yu, J.J. & Shen, F. The IL-17 cytokine family. Vitam. Horm. 74, 255–282 (2006).
Jurgens, M. et al. Disease activity, ANCA, and IL23R genotype status determine early response to infliximab in patients with ulcerative colitis. Am. J. Gastroenterol. 105, 1811–1819 (2010).
Tillack, C. et al. Anti-TNF antibody-induced psoriasiform skin lesions in patients with inflammatory bowel disease are characterised by interferon-gamma-expressing Th1 cells and IL-17A/IL-22-expressing Th17 cells and respond to anti-IL-12/IL-23 antibody treatment. Gut 63, 567–577 (2014).
Acknowledgements
This paper is part of the degree thesis of Matthias Friedrich. We thank Andrea Bedynek (Department of Medicine II—Grosshadern, Ludwig-Maximilians-University (LMU) Munich, Germany), Brigitte Hackl (Clinic for Preventive Dentistry and Parodontology, LMU Munich, Germany), Heike Borth (Walther-Straub-Institute, LMU Munich, Germany), Sonja Kresse (Department of Dermatology and Allergy, LMU Munich, Germany), and Julian Maurer (Department of Medicine II—Grosshadern, Ludwig-Maximilians-University (LMU) Munich, Germany) for excellent technical support. We thank Rajita Pappu (Department of Immunology, Genentech, CA) for very helpful discussions. S.B. was supported by grants from the Else-Kröner-Fresenius-Stiftung (Else Kröner-Exzellenzstipendium 2010_EKES.32) and the Deutsche Forschungsgemeinschaft (DFG; BR 1912/6-1). J.S. was supported by the Deutsche Forschungsgemeinschaft (Scha 979/5-1) and Else-Kröner-Fresenius Stiftung (2012_A119).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Additional information
Author contributions
M.F.: study design, experiments, and manuscript writing; J.D.: study design and experiments; J.S.: experiments and funding; C.J.A. study design and experiments; and S.B.: study design, patient recruitment, manuscript writing, and funding.
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
PowerPoint slides
Rights and permissions
About this article
Cite this article
Friedrich, M., Diegelmann, J., Schauber, J. et al. Intestinal neuroendocrine cells and goblet cells are mediators of IL-17A-amplified epithelial IL-17C production in human inflammatory bowel disease. Mucosal Immunol 8, 943–958 (2015). https://doi.org/10.1038/mi.2014.124
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2014.124
This article is cited by
-
IL-17C is a driver of damaging inflammation during Neisseria gonorrhoeae infection of human Fallopian tube
Nature Communications (2024)
-
Lactobacillus paracasei R3 protects against dextran sulfate sodium (DSS)-induced colitis in mice via regulating Th17/Treg cell balance
Journal of Translational Medicine (2021)
-
A defective interleukin-17 receptor A1 causes weight loss and intestinal metabolism-related gene downregulation in Japanese medaka, Oryzias latipes
Scientific Reports (2021)
-
Toll-like receptor 5-mediated IL-17C expression in intestinal epithelial cells enhances epithelial host defense against F4+ ETEC infection
Veterinary Research (2019)
-
Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity
Mucosal Immunology (2018)