Abstract
Bacterial immunoglobulin A1 (IgA1) proteases may sabotage the protective effects of IgA. In vitro, both exogenous and endogenously produced IgA1 protease inhibited phagocytic killing of Streptococcus pneumoniae by capsule-specific IgA1 human monoclonal antibodies (hMAbs) but not IgA2. These IgA1 proteases cleaved and reduced binding of the the effector Fcα1 heavy chain but not the antigen-binding F(ab)/light chain to pneumococcal surfaces. In vivo, IgA1 protease-resistant IgA2, but not IgA1 protease-sensitive IgA1, supported 60% survival in mice infected with wild-type S. pneumoniae. IgA1 hMAbs protected mice against IgA1 protease-deficient but not -producing pneumococci. Parallel mouse sera with human IgA2 showed more efficient complement-mediated reductions in pneumococci with neutrophils than did IgA1, particularly with protease-producing organisms. After natural human pneumococcal bacteremia, purified serum IgG inhibited IgA1 protease activity in 7 of 11 patients (64%). These observations provide the first evidence in vivo that IgA1 protease can circumvent killing of S. pneumoniae by human IgA. Acquisition of IgA1 protease-neutralizing IgG after infection directs attention to IgA1 protease both as a determinant of successful colonization and infection and as a potential vaccine candidate.
Similar content being viewed by others
Introduction
Although Streptococcus pneumoniae infections begin as asymptomatic nasopharyngeal colonization, this organism is the most common bacterial cause of community-acquired pneumonia (≈20–30% of cases), purulent otitis media (10–25%), and sinusitis.1, 2, 3, 4 As an invasive pathogen, the pneumococcus is the leading cause of bacteremic pneumonia (≈65%) and bacterial meningitis (>30%),5 with ⩾20,000 deaths per year in the United States6 and up to a million deaths worldwide.7 The functional interplay between bacterial virulence factors and host immune responses likely determines whether S. pneumoniae causes asymptomatic colonization, isolated mucosal infection, or invasive disease.
In both the blood and upper respiratory tract, immunoglobulin A1 (IgA1) comprises ≈90% of total IgA, whereas IgA2 accounts for only ≈10%.8, 9 Human IgA1 is cleaved by highly host species-specific proteases produced by S. pneumoniae and several other invasive mucosal pathogens (e.g., Neisseria species and Haemophilus influenzae). Cleavage results in dissociation of the effector function of the Fc fragment of bound IgA1 from its antigen-binding Fab fragments,10, 11, 12, 13, 14 a process shown to promote adherence of S. pneumoniae to respiratory epithelial cells in vitro.15 IgA1 protease had been proposed,16, 17, 18, 19 and, more recently, shown in vitro to abrogate the protective effects of human IgA1,20 effects that include complement-dependent killing of the organism by phagocytes. However, because of the species- and subclass-specificity of the protease and the previous unavailability of purified IgA1 and IgA2 of comparable antigen-specificities and functional activity from humans,21, 22, 23 the contribution of IgA1 proteases to bacterial survival in vivo has not been demonstrated. Thus, we investigated the ability of IgA1 protease to modify killing of S. pneumoniae by using novel IgA1 and IgA2 human monoclonal antibodies (hMAbs) specific for the pneumococcal capsule, isogenic wild-type and IgA1 protease-deficient organisms, and both in vitro and in vivo assay systems.
Results
IgA1 protease inhibits IgA-dependent killing of S. pneumoniae in vitro
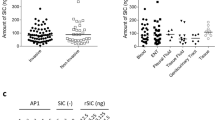
Pneumococcal capsule–specific hMAbs of the IgA1 and IgA2 subclasses can kill wild-type S. pneumoniae in vitro (reduce the number of bacterial colony-forming units (CFU)) in the presence of complement and phagocytic cells.20 We determined the ability of IgA1 protease, which cleaves the capsule-binding variable region of IgA1 from the phagocyte-binding constant region, to disrupt killing by these antibodies. Indeed, intact capsule-specific IgA1 hMAbs supported dose-dependent killing of S. pneumoniae (Figure 1a). However, pretreatment of the IgA1 hMAbs with IgA1 protease (available in recombinant form from H. influenzae Rd) eliminated this IgA-mediated killing of S. pneumoniae (67±6% vs. 10±4% kill with and without IgA1 protease preincubation, respectively; P<0.01). By contrast, the protease had no effect on killing supported by the capsule-specific IgA2 hMAbs (Figure 1b). IgA1 protease cleaved the heavy chain of IgA1, but not of IgA2, into lower molecular weight fragments (inserts; Figures 1a, respectively), although a fraction of the heavy chain remained intact. The protease had comparable inhibitory effects on killing of type 8 organisms with a type 8–specific IgA1 hMAbs (not shown). The killing resulted primarily from the opsonophagocytic activity of capsule-specific IgA and complement with neutrophils. However, a small proportion of the decrease in CFUs in the killing results (3–5% at 75 ng ml−1 IgA1, 11–15% at 225 ng ml−1) can be ascribed to agglutination of the organisms by polymeric IgA alone rather than phagocytosis (Fasching et al.20 and Supplementary Figure S1).
Immunoglobulin A1 (IgA1) protease inhibits killing of Streptococcus pneumoniae type 2 by human monoclonal antibodies (hMAbs). (a) Type 2 capsule-specific human IgA1 (hMAb 2A02) and (b) IgA2 hMAbs (MAb 2A01) (b) were incubated overnight with exogenous purified recombinant IgA1 protease from Haemophilus influenzae Rd (digested) or phosphate-buffered saline (undigested). After protease was removed by binding the IgA1 protease 6 × His tag on a nickel column, killing of S. pneumoniae type 2 (strain 6302; ATCC) with IgA1 or IgA2 hMAbs was performed in the presence of 10% complement and human neutrophils (polymorphonuclear neutrophils (PMN): bacteria ratio 500:1), as described in Methods. Inserts: Cleavage of human IgA1 hMAbs (a) but not IgA2 hMAbs (b) by recombinant IgA1 protease (3 μg). Products resolved on the 12% reducing SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) gel include intact heavy chain and cleavage fragments (black arrows) and light chain (white arrows). (c, d) Pneumococcal killing experiments were performed to determine the effects of untreated IgA1 and IgA2 MAbs against IgA1 protease-positive wild-type S. pneumoniae (P210) and the isogenic IgA1 protease-negative mutant (P354). In these experiments, the bacteria themselves were the only source of IgA1 protease. Dotted lines in c and d represent the concentration of hMAbs required for 50% kill with complement and PMNs. Open circles (○) represent IgA1 protease exposure (exogenous, a and b; or endogenous, c and d) and closed circles (●) represent no IgA1 protease exposure. PPS, pneumococcal capsular polysaccharides.
Consistent with the effects of preincubation of IgA1 with exogenous IgA1 protease, endogenous IgA1 protease produced directly by S. pneumoniae also inhibited such killing. Killing of protease-producing organisms required ≈fivefold more IgA1 hMAbs than did killing of isogenic IgA1 protease–deficient mutants (for 50% kill, 70 vs. 15 ng ml−1, respectively) (Figure 1c). By contrast, the IgA2 hMAbs supported killing of both organisms with equal efficiency (Figure 1d). After overnight incubation, supernatant fluid from the wild-type but not the protease-deficient S. pneumoniae cleaved the heavy chain of IgA1, but not IgA2 (not shown). Thus, IgA1 protease from H. influenzae, as applied exogenously, and from S. pneumoniae, produced endogenously, diminished the ability of human IgA1 to support killing of S. pneumoniae by phagocytes with complement.
Binding of IgA to S. pneumoniae in vitro
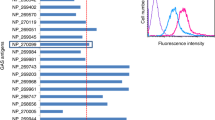
We considered whether IgA1 protease inhibited binding of the heavy chain alone or the whole IgA1 antibody to the organism. Preincubation of capsule-specific IgA1 hMAbs with exogenous recombinant IgA1 protease significantly reduced the amount of IgA1 heavy chain bound to S. pneumoniae to 9% of that with intact IgA1 as detected by IgA1-specific detector antibody (Figure 2a). Similarly, exposure of the IgA1 hMAbs to pneumococcal IgA1 protease produced endogenously over 90 min by the organism during incubation also decreased detectable bound IgA1 by 43%. However, cleavage of IgA1 by the protease did not affect binding of the light chain and likely the F(ab) fragment to the organism (Figure 2c).
Immunoglobulin A1 (IgA1) protease reduces IgA1 Fcα bound to Streptococcus pneumoniae. Detection of serotype-specific human IgA1 hMAbs (human monoclonal antibodies) bound to serotype 2 organisms (P210). (a) Organisms were exposed on ice for 30 min to intact hMAbs (black and striped bars) or hMAbs predigested with exogenous purified IgA1protease from Haemophilus influenzae Rd (gray bar) and washed. After 5 min at 37 °C (black and gray bars) or 90-min exposure of intact hMAbs to endogenous protease produced by the S. pneumoniae organisms (striped bar), residual-bound IgA1 was measured on washed bacteria stained with phycoerythrin-labeled mouse anti-human IgA1 by flow cytometry. Values for mean fluorescence intensity (MFI)±s.d. with intact MAb in six experiments are considered as 100%. P<0.001 for exogenous and P<0.02 for endogenous pneumococcal IgA1 protease compared with intact MAb. (b) Ninety-minute incubation of living wild-type S. pneumoniae (P210) resulted in loss of IgA1 (striped bar), but we observed no such loss of bound IgA1 from fluorescein isothyocyanate–tagged dead organisms alone (black bar) or dead organisms that were washed, followed by addition of an equal number of unopsonized untagged wild-type live organisms (gray bar). The percentage binding of IgA1 (Fcα), as determined by the MFI at 90 min, was determined by flow cytometry, as in a. Data are shown as the mean±s.d. for four experiments. (c) Protease-deficient S. pneumoniae (strain P354) are incubated with intact or protease-treated IgA1 or IgA2 are incubated for 90 min, washed, and surface Fcα heavy chains and κ light chains are detectedy by flow cytometry. IgA1 protease decreases the amount of binding of IgA1 Fcα to S. pneumoniae (P=0.007) but neither the κ light chain binding nor the Fcα, which is resistant to the enzyme. Results are shown as mean of maximum binding (MFI)±s.d. for two experiments.
We next considered whether the effects of the endogenously produced protease on IgA1 were due to enzymatic activity expressed on the bacterial cell surface or released into solution. When specific monoclonal IgA1 was incubated with live wild-type S. pneumoniae for 90 min, less than half the IgA1 heavy chain was detected on the bacterial surface with the anti-IgA1 detector antibody compared with that bound to heat-killed organisms (Figure 2b). However, incubation of IgA1 hMAbs with heat-killed organisms caused no such decrement in bound antibody, suggesting that the protease is heat-labile or the activity required active production. Moreover, co-incubation of IgA1-opsonized heat-killed fluorescein isothyocyanate (FITC)-tagged organisms with an equal number of untagged and unopsonized live IgA1 protease–producing organisms over the same period also resulted in no decrement in IgA1 detection on the heat-killed bacterial surface. Thus, the proteolytic effects of IgA1 protease on capsule-specific IgA1 bound to bacteria appears to occur primarily on IgA1 bound to the cell surface of living bacteria and are not due to effects of release of the enzyme into the fluid phase of the culture medium.15 Consistent with this observation, IgA1 protease is anchored to the cell surface and not actively secreted.24 However, IgA1 bound to S. pneumoniae can be cleaved by soluble IgA1 protease added to the media so that bound antibody is not protected from cleavage by exogenous enzyme (data not shown). Whether live organisms can produce enough, and release sufficient, IgA1 protease to cleave the antibody on neighboring organisms in vivo has not been proven to date.
IgA1 protease eliminates IgA1-mediated protection against S. pneumoniae in vivo
We compared the ability of the two capsule-specific hMAbs, each of which supported killing of S. pneumoniae in vitro, to provide passive protection against fatal infection in vivo. After intraperitoneal administration of the protease-resistant IgA2 hMAbs, 81% of mice given intranasal challenge of wild-type S. pneumoniae survived at 72 h compared with 10% of those pretreated with IgA1 hMAbs or 17% of those given phosphate-buffered saline control (P<0.0001; Figure 3a). However, levels of the two hMAbs achieved in mouse sera after passive immunization were comparable over time (Figure 3b). No human IgA was detected in the nares. These IgA subclass–related differences in survival were associated with differences in the ability of sera from mice passively immunized with IgA1 and IgA2 hMAbs to support complement-dependent killing by phagocytes in vitro (Figure 3c). The IgA1 hMAbs in mouse sera from the earliest time point (4 h), which contained the highest hMAb level, killed the IgA1 protease–producing wild-type organism effectively (right column pair; black bar). However, this activity declined over time as antibody levels declined (right column pair; grey bar). By contrast, these same sera consistently supported killing of an isogenic IgA1 protease–deficient mutant at both time points examined (second column pair). With the protease-resistant IgA2 hMAbs, killing of both organisms was comparable at each time and antibody concentration (first and third column pairs). In addition, a serotype-specific IgG1 MAb also showed similar killing of both wild-type and mutant organisms (not shown). The heat-inactivated mouse serum had no activity against the organisms. Thus, the pneumococcal protease appears to specifically target human IgA1 and not other aspects of phagocyte-dependent killing.
Protection of mice against fatal infection with serotype 2 wild-type Streptococcus pneumoniae by human immunoglobulin A (IgA) monoclonal antibodies. (a) Survival of mice pre-treated intraperitoneally with 2 μg human IgA1 (open circle, ○; n=36 mice), IgA2 (closed circle, ●; n=37) human monoclonal antibodies (hMAbs), or buffer (Δ; n=53) 4 h before intranasal challenge with wild-type S. pneumoniae (P210; P<0.00001 for IgA2 vs. both the other groups). (b) We measured changes in the concentrations of specific IgA1 and IgA2 hMAbs in pooled mouse serum by enzyme-linked immunosorbent assay in control mice (four per group) over time after passive immunization. (c) We tested the ability of sera containing IgA1 or IgA2 hMAbs from these animals obtained over time at 4 and 24 h to support killing by human neutrophils of IgA1-producing wild-type S. pneumoniae (positive; strain P210) or its isogenic IgA1 protease-deficient mutant (negative; strain P354) in the presence of 10% baby rabbit complement (c). Data are shown as mean±s.d. for two experiments using pooled sera from four mice at each time point with each condition. PBS, phosphate-buffered saline.
Finally, we considered the possibility that differences in the activity of IgA1 and IgA2 hMAbs accounted for the differential protection afforded by the two hMAbs, rather than their susceptibility to cleavage by IgA1 protease or in the virulence of the organisms. Indeed, the IgA2 hMAbs supported killing more efficiently than did the IgA1 hMAbs in vitro (Figures 1a). However, passive immunization of mice with the IgA1 hMAbs was associated with 50% survival at 72 h when the mice were infected with the protease-deficient mutant, but only 10% when infected with the isogenic protease-producing parent (P<0.005; Figure 4). Moreover, in the absence of antibody, the virulence of these isogenic organisms was not significantly different for the protease-deficient vs. wild-type strains, as reflected by the lethal dose50 (LD50: 3 × 106 CFU and 1 × 107 CFU, respectively) by Reed–Muench analysis25 with intranasal infection. Moreover, the in vitro growth curves of the two organisms were similar (not shown). Therefore, production of IgA1 protease by the organisms, and the susceptibility of the hMAbs to protease cleavage, was closely associated with the ability of the capsule-specific IgA antibodies to protect against this encapsulated pathogen in vitro and in vivo.
Effect of Immunoglobulin A1 (IgA1) protease on protection by human monoclonal antibody IgA1. Mice were passively immunized as described in the legend of Figure 3 and challenged intranasally with IgA1 protease-producing wild-type Streptococcus pneumoniae type 2 (Strain P210; n=15 mice) or its IgA1 protease-deficient mutant (Strain P354; n=15 mice).
Human immune IgG inhibits IgA1 protease
Preincubation of IgA1 with pneumococcal IgA1 protease substantially reduced detection of the hinge region with a hinge-specific murine monoclonal antibody by immunoblot (Figure 5a; controls). Using this detection system for protease activity, purified IgG from convalescent sera from two representative patients obtained 1 month after resolution of pneumococcal bacteremia inhibited the protease activity, allowing detection of the hinge region. Acute sera from these patients showed no inhibition of protease activity, so the hinge region was not detected.
Inhibition of Immunoglobulin A1 (IgA1) protease activity by human serum IgG. (a) Protection of the IgA1 heavy chain hinge region (arrow) from cleavage by IgA1 protease was determined by immunoblotting with murine anti-human IgA1 monoclonal antibodies (MAb). Purified serum IgA was incubated without (−-) or with (+) IgA1 protease (partially purified from lysed Streptococcus pneumoniae type 14) alone or in the presence of purified IgG from acute (Ac) and convalescent (Conv) sera from two patients with pneumococcal bacteremia drawn within 2 days of and 28 days after initial presentation to the hospital, respectively. (b) The reciprocal protease inhibitory titers of purified serum IgG from 11 adult patients with pneumococcal bacteremia include those from 7 patients whose titers rose (closed triangle, ▴) in convalescent sera and 4 whose titers did not (open circle, ○).
Among 11 patients surviving bacteremic pneumococcal pneumonia and tested within 2–7 days of the onset of symptoms, two (18%) (both capsular serotype 14) had detectable IgA1 protease–inhibiting activity in acute sera drawn within 3 days of admission (Figure 5b). In convalescent sera drawn 3–4 weeks after infection, 7 of 11 (64%) patients, including the 2 with detectable activity in acute sera, generated significantly increased titers of IgA1 protease–inhibiting IgG. Median titers of IgA1 protease–neutralizing activity rose from 1:3 to 1:20 in acute and convalescent sera, respectively. The serum IgG concentrations required for inhibition of IgA1 protease activity ranged from 1.2 to 10 μg ml−1 (total serum IgG levels were 8–12 mg ml−1). Among these samples, neutralizing activity was largely independent of the serotype of the infecting organisms. Four of these patients were infected with serotype 14 S. pneumoniae isolated from blood during their acute bacteremia, and one each had types 1, 4 and 8. These data suggest neutralizing antibodies elicited during natural infection may inhibit the proteolytic activity of bacterial IgA1 proteases. The inability to generate neutralizing activity in four patients may have been related to antigenic differences between the proteases26 or an inability of these patients to respond.
Discussion
By exploiting the functional activity of novel serotype-matched human IgA1 and IgA2 hMAbs, we show that IgA1 protease compromises the ability of the capsule-specific IgA1 to provide protection against S. pneumoniae both in vitro and in vivo. For decades, IgA1 proteases have been proposed to enhance the pathogenicity of invasive mucosal bacteria.16, 17, 27 Different bacterial species express functionally similar proteases that cleave the human IgA1 hinge region after a sequence of specific proline residues, but the proteases have completely different catalytic mechanisms.17 This apparent “convergent evolution” of the enzyme activity among the many different mucosal bacterial pathogens17 suggests that the activity promotes bacterial survival. However, there has been no direct demonstration of the importance of bacterial IgA1 proteases in vivo, because the native IgA of animals used for these experiments is not a substrate for the IgA proteases. Experimental human urethral infection was successful with an IgA1 protease mutant of Neisseria gonorrhoeae, but IgA1 is not the predominant IgA subclass at this site.28 In addition, the mechanism(s) by which the protease accomplishes a benefit to the bacteria in vivo had not been demonstrated.
The majority of bacteria producing IgA-cleaving proteases colonize the oral and upper respiratory mucosa where IgA1 predominates.29, 30 Thus, IgA1 protease from S. pneumoniae may contribute to the bacteria’s ability to colonize and invade by impairing the function of IgA1. Consistent with this hypothesis, we have shown that pneumococcal capsule–specific IgA1 enhances, rather than inhibits, adherence of encapsulated S. pneumoniae to human respiratory epithelium in the presence of IgA1 protease.15
Although such processes may contribute to colonization, the protease may also facilitate the development of clinical disease by other mechanisms. For example, levels of IgA1 protease activity from H. influenzae are increased in strains causing tissue injury and disease compared with those causing colonization alone.31 The assays we employed in this study, killing by neutrophils and animal protection by systemic antibody following mucosal challenge, may be more directly related to the ability of IgA to protect against disease rather than colonization. We showed that capsule-specific human MAbs of the protease-resistant IgA2 subclass are equal in supporting the killing of wild-type and protease-deficient mutant organisms, a result that suggests that pneumococcal IgA protease does not appear to impair the phagocytic function of the neutrophils themselves in these settings. In related studies, we have also not identified significant differences in the virulence for mice of isogenic wild-type or protease-deficient mutants of S. pneumoniae serotype 2.32 However, that IgA1 protease did influence survival of infected mice in other reports33, 34, 35 suggests that such differences may depend on the inoculum (less effect at higher inoculum), the bacterial serotype (type 4 and 19F vs. type 2), the strain of mice (out bred MF1 and inbred BALB/c vs. out bred specific pathogen-free Swiss mice), the clinical model (pneumonia vs. bacteremia), and perhaps the specific molecular deletion, each of which differed from our studies. That mice do not produce IgA that is susceptible to IgA1 protease suggests other functions for the protein in bacterial pathogenesis.
The reduction in killing activity by IgA1 following the cleavage of its heavy polypeptide chain by IgA protease is likely due to the known dissociation of the effector domains of the antibody, unlinking the Fcα CH2/3 fragments that are essential for binding to Fcα receptors (CD89) on neutrophils and alveolar macrophages from the bacterial-binding Fab domains.36, 37, 38 These Fcα CH2/3 domains have been associated with complement activity in both the polymeric and secretory forms of IgA.39, 40 In addition, protease cleavage yields Fab domains that appear to continue to bind to antigens on the bacterial surface. However, the monomeric Fab binding would not foster the formation of aggregates that would support bacterial clearance, which depends on the Fc region of the antibody.41 Although we have focused on killing and invasive infection, the protease may well also limit agglutination, enhance adherence15 and potentially killing at mucosal sites, activities currently under investigation.
Unlike the IgA1 proteases of Gram-negative pathogenic microorganisms that are secreted into the medium, the protease produced by S. pneumoniae does not appear to be actively secreted but apparently reaches the medium only when cell lysis has taken place. The IgA1 protease of S. pneumoniae is predominantly bacterial cell surface–associated through an unusual sortase A-dependent, non-covalent mechanism of attachment24 Our data confirm recent studies that the activity of the protease that is retained at the bacterial surface may have substantial impact on specific pneumococcal capsule–specific antibodies that are directly bound to the pathogen. By contrast, a protease secreted in free form into the medium may be expected to have a more generalized effect on IgA in the mucosal environment, thus facilitating IgA1 evasion by other microbes that are competing for the same niche.9, 15, 32
We demonstrate that the activity of IgA1 protease can be inhibited by serum IgG antibodies arising during convalescence from invasive pneumococcal disease. This neutralization of proteolytic activity aligns with earlier observations that antibodies that bind42 and block activity of the proteases of Neisseriae, H. influenzae, and other species43, 44, 45 can be found in serum and secretions, such as breast milk and nasal fluids. Romanello et al.46 showed that sera from patients with invasive pneumococcal disease recognized recombinant pneumococcal IgA protease proteins, but these investigators did not report that these antibodies inhibited the enzyme activity. The impact of our results may be limited by the antigenic diversity of IgA1 proteases that has been found among clinical pneumococcal isolates, both between and within serotypes.26 Ongoing functional and sequencing data with these strains will determine whether the proteolytic regions and neutralizing domains are most well conserved.47 The immunological consequences of protease heterogeneity can be determined with greater numbers of patients and a more thorough understanding of proteolytic motifs and inhibitory epitopes. Nevertheless, our results highlight the potential of IgA1 protease as a vaccine candidate that may limit colonization on mucosal surfaces served by secretory IgA1 antibodies and that may reduce the incidence of invasive disease and local mucosal infections caused by this important bacterial pathogen.
In summary, IgA1 proteases produced by S. pneumoniae, and other invasive mucosal pathogens, may enable the bacteria to evade killing by capsule-specific IgA by sabotaging the effector function of the specific humoral immune response. This proteolytic activity of the pneumococcal enzyme appears to be primarily cell-associated and directed to pathogen-specific IgA antibodies bound to the organism. However, this activity is only one element of the host–pathogen interaction in which the pathogen attempts to establish a niche in the context of robust host defense. Following colonization, S. pneumoniae can elicit a local and/or systemic IgA1 response to the bacterial surface.30, 48 As demonstrated here, the pathogen produces IgA1 protease that limits the protective effects of this antibody. In turn, the host then generates a specific antibody response in blood (or mucosa) that neutralizes the proteolytic activity of the enzyme. The interplay between bacterial virulence factors and the host’s successive ability to circumvent them likely determines the outcome of these common and often serious infections.
Methods
S. pneumoniae strains. Strains included wild-type S. pneumoniae isolates of serotypes 2 (ATCC 6302) and 8 (ATCC 6308) (American Tissue Culture Collection; Mannassas, VA), and paired isogenic serotype 2 wild-type and insertion–deletion IgA1 protease mutant organisms, which yielded a nonfunctional truncated IgA1 protease gene product (strains P210 and P354, respectively).32
IgA1 protease. We used recombinant IgA1 protease from an unencapsulated H. influenzae strain Rd mutant with a nickel-binding 6 × His epitope replacement44 (recombinant pneumococcal IgA1 protease not available), and partially purified pneumococcal IgA1 protease, derived by differential precipitation (30 and 60% ammonium sulfate) of supernatants of log-phase S. pneumoniae type 14 (ATCC 6314) in Todd–Hewitt broth.49 Enzymatic activity was confirmed using silver-stained denaturing polyacrylamide gel electrophoresis (12%) of IgA1 and IgA2 hMAb fragments before and after overnight incubation with IgA1 protease.
hMAbs to pneumococcal capsular polysaccharides. hMAbs specific for S. pneumoniae serotypes 2 (IgA1 (2A02), IgA2 (2A01), IgG1 (2G01)), and serotype 8 (IgA1 (8A01)) were purified to >98.5% purity from B cells of recently immunized healthy adults that had been fused with the K6H6/B5 mouse–human heteromyeloma and cloned 3 × by limiting dilution, as described.15, 18, 50, 51 Consistent with results with capsule-specific IgA in serum, the majority of the MAb IgA is in the polymeric molecular form.18, 20, 52
In vitro killing assay. Log-phase S. pneumoniae serotypes 2 (ATCC 6302, P210, P354) or 8 (ATCC 6308) were incubated with hMAbs for 30 or 120 min, then 1 h with baby rabbit complement (10%) and human neutrophils (500:1, neutrophil:bacteria) with shaking.18 CFU were determined from serial dilutions on blood agar plates. Percentage of kill was determined by: (1−(CFU in wells with MAb per CFU in wells without MAb)) × 100.
Murine protection model. In protocols approved by the animal research committee, after inhaled isoflurane anaesthesia, specific pathogen-free Swiss out bred mice in a pathogen-free barrier facility were inoculated intranasally with 2 × 107 log-phase CFU bacterial suspension (≈2 × LD50) 4 h after an intraperitoneal dose of human capsule–specific hMAbs (2 μg) or saline placebo. Survival was recorded every 8 h until killing at 72 h.53
IgA-bacterial binding assays. S. pneumoniae serotype 2 strains (ATCC 6302 and P210) were incubated 30 min with capsule-specific IgA1 hMAbs that had or had not been digested overnight by recombinant IgA1 protease. Alternatively, an IgA1 protease–producing wild-type strain (P210) was incubated with intact capsule-specific IgA1 hMAbs for 30 min, washed, and incubated for 90 min. Bacteria were stained with mouse anti-human IgA1-biotin (Southern Biotechnology Associates, Birmingham, AL), followed by streptavidin (SA)-allophycocyanin (BD Pharmingen, San Diego, CA). The mean fluorescent intensities of the bacteria, identified by forward and side scatter, were compared (FACSVantage cytometer; Becton Dickinson, Mansfield, MA) using control antibodies matched for species, source, isotype, and fluorescent label and compensation control beads. The mouse anti-IgA1 neither recognized bound IgG or any variable region of the immunoglobulin heavy chain regions that might be shared by IgA and IgG antibodies nor its associated κ or λ light chains (data not shown).
To consider whether the cleavage of the IgA1 occurred on the bacterial surface or in the fluid phase, live unlabeled or dead FITC-tagged wild-type S. pneumoniae type 2 were opsonized with specific IgA1 hMAbs for 30 min at 37 °C and washed. Three populations of bacteria (opsonized live, opsonized dead, or opsonized FITC-tagged dead+unopsonized live) were incubated for an additional 90 min and stained. Dead bacteria were segregated by gating on FITC+ events. Mean fluorescent intensities for bound IgA1 were compared between groups.
Inhibition of IgA1 protease cleavage by human IgG. Sera were obtained with protocols approved by the relevant institutional review boards at admission to the Minneapolis Veterans Affairs Medical Center and from convalescent sera 3–4 weeks later from 11 adult patients with bacteremic pneumococcal pneumonia (blood stream infection) and symptoms for 2–7 days but without HIV-1 infection or obvious humoral immune defects. We purified IgG (purity >98%) from IgA-depleted acute sera exactly as described from our laboratory.18 Pneumococcal protease was preincubated with purified IgG dilutions (initial 20 μg IgG ml−1) for 15 min at 37 °C, then incubated with 2.5 μg of purified control serum IgA (>98% pure) for 2 h. Inhibition of cleavage was determined by the percentage of loss of heavy chain by densitometry on reducing 10% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and by detection or loss of the IgA1 hinge with mouse anti-human IgA1-biotin by immunoblot.
Statistics. Survival of animals in the treatment groups was compared using Cox regression analysis, and if statistically significant differences were found, survival curves were compared by log-rank test. Statistical comparisons were calculated using SPSS for Windows 6.1 (IBM, Chicago, IL).
References
Dagan, R., Engelhard, D. & Piccard, E. Epidemiology of invasive childhood pneumococcal infections in Israel: the Israeli Pediatric Bacteremia and Meningitis Group. JAMA 268, 3328–3332 (1992).
Bruyn, G.A.W., van der Meer, J.W.M., Hermans, J. & Knoppert, W. Pneumococcal bacteremia in adults over a 10-year period at University Hospital, Leiden. Rev. Infect. Dis. 10, 446–450 (1988).
Jetté, L.P. & Lamothe, F. Surveillance of invasive Streptococcus pneumoniae infection in Quebec, Canada, from 1984 to 1986: serotype distribution, antimicrobial susceptibility, and clinical characteristics. J. Clin. Microbiol. 27, 1–5 (1989).
Kuikka, A., Syrjanen, J., Renkonen, O.V. & Valtonen, V.V. Pneumococcal bacteraemia during a recent decade. J. Infect. 24, 157–168 (1992).
Thigpen, M.C. et al. Bacterial meningitis in the United States, 1998–2007. N. Engl. J. Med. 364, 2016–2025 (2011).
Centers for Disease Control and Prevention. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR 46, 1–24 (1997).
O'Brien, K.L. et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902 (2009).
Brandtzaeg, P. Regionalized immune function of tonsils and adenoids. Immunol. Today 20, 383–384 (1999).
Kirkeby, L., Rasmussen, T.T., Reinholdt, J. & Kilian, M. Immunoglobulins in nasal secretions of healthy humans: structural integrity of secretory immunoglobulin A1 (IgA1) and occurrence of neutralizing antibodies to IgA1 proteases of nasal bacteria. Clin. Diagn. Lab. Immunol. 7, 31–39 (2000).
Male, C.J. Immunoglobulin A1 protease production by Haemophilus influenzae and Streptococcus pneumoniae. Infect. Immun. 26, 254–261 (1979).
Kilian, M., Mestecky, J. & Schrohenloher, R.E. Pathogenic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. Infect. Immun. 26, 143–149 (1979).
Kilian, M., Mestecky, J., Kulhavy, R., Tomana, M. & Butler, W.T. IgA1 proteases from Haemophilus influenzae, Streptococcus pneumoniae, Neisseria meningitidis, and Streptococcus sanguis: comparative immunochemical studies. J. Immunol. 124, 2596–2600 (1980).
Mulks, M.H. & Plaut, A.G. IgA protease production as a characteristic distinguishing pathogenic from harmless neisseriaceae. N. Engl. J. Med. 299, 973–976 (1978).
Plaut, A.G. The IgA1 proteases of pathogenic bacteria. Annu. Rev. Microbiol. 37, 603–622 (1983).
Weiser, J.N. et al. Antibody-enhanced pneumococcal adherence requires IgA1 protease. Proc. Natl. Acad. Sci. USA 100, 4215–4220 (2003).
Kilian, M., Reinholdt, J., Lomholt, H., Poulsen, K. & Frandsen, E.V. Biological significance of IgA1 proteases in bacterial colonization and pathogenesis: critical evaluation of experimental evidence. APMIS 104, 321–338 (1996).
Kilian, M., Mestecky, J. & Russell, M.W. Defense mechanisms involving Fc-dependent functions of immunoglobulin A and their subversion by bacterial immunoglobulin A proteases. Microbiol. Rev. 52, 296–303 (1988).
Janoff, E.N. et al. Killing of Streptococcus pneumoniae by capsular polysaccharide-specific polymeric IgA, complement, and phagocytes. J. Clin. Invest. 104, 1139–1147 (1999).
Finn, A. et al. Induction of functional secretory IgA responses in breast milk by pneumococcal capsular polysaccharides. J. Infect. Dis. 186, 1422–1429 (2002).
Fasching, C.E. et al. Impact of the molecular form of IgA on its functional activity in defense against Streptococcus pneumoniae. Infect. Immun. 75, 1801–1810 (2007).
Steinitz, M., Tamir, S., Ferne, M. & Goldfarb, A. A protective human monoclonal IgA antibody produced in vitro: anti-pneumococcal antibody engendered by Epstein-Barr virus-immortalized cell line. Eur. J. Immunol. 16, 187–193 (1986).
Baxendale, H.E. et al. Immunogenetic analysis of the immune response to pneumococcal polysaccharide. Eur. J. Immunol. 30, 1214–1223 (2000).
Zhong, Z., Burns, T., Chang, Q., Carroll, M. & Pirofski, L. Molecular and functional characteristics of a protective human monoclonal antibody to serotype 8 Streptococcus pneumoniae capsular polysaccharide. Infect. Immun. 67, 4119–4127 (1999).
Bender, M.H. & Weiser, J.N. The atypical amino-terminal LPNTG-containing domain of the pneumococcal human IgA1-specific protease is required for proper enzyme localization and function. Mol. Microbiol. 61, 526–543 (2006).
Reed, L.J. & Muench, H. A simple method of estimating fifty percent endpoints. Am J. Hyg. 27, 493–497 (1938).
Lomholt, H. Evidence of recombination and an antigenically diverse immunoglobulin A1 protease among strains of Streptococcus pneumoniae. Infect. Immun. 63, 4238–4243 (1995).
Plaut, A.G., Gilbert, J.V., Artenstein, M.S. & Capra, J.D. Neisseria gonorrhoeae and Neisseria meningitidis: extracellular enzyme cleaves human immunoglobulin A. Science 190, 1103–1105 (1975).
Johannsen, D.B., Johnston, D.M., Koymen, H.O., Cohen, M.S. & Cannon, J.G. A Neisseria gonorrhoeae immunoglobulin A1 protease mutant is infectious in the human challenge model of urethral infection. Infect. Immun. 67, 3009–3013 (1999).
Kilian, M., Thomsen, B., Petersen, T.E. & Bleeg, H.S. Occurrence and nature of bacterial IgA proteases. Ann. NY Acad. Sci. 409, 612–624 (1983).
Opstad, N.L. et al. Impact of Streptococcus pneumoniae bacteremia and human immunodeficiency virus type 1 on oral mucosal immunity. J. Infect. Dis 172, 566–570 (1995).
Vitovski, S., Dunkin, K.T., Howard, A.J. & Sayers, J.R. Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels. JAMA 287, 1699–1705 (2002).
Wani, J.H., Gilbert, J.V., Plaut, A.G. & Weiser, J.N. Identification, cloning, and sequencing of the immunoglobulin A1 protease gene of Streptococcus pneumoniae. Infect. Immun. 64, 3967–3974 (1996).
Chiavolini, D., Memmi, G., Maggi, T., Ianelli, F., Pozzi, G. & Oggioni, M.R. The three extra-cellular zinc metalloproteinases of Streptococcus pneumoniae have a different impact on virulence in mice. BMC Microbiol. 3, 14 (2003).
Polissi, A. et al. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66, 5620–5629 (1998).
Oggioni, M.R. et al. Pneumococcal zinc metalloproteinase ZmpC cleaves human matrix metalloproteinase 9 and is a virulence factor in experimental pneumonia. Mol. Microbiol. 49, 795–805 (2003).
Shen, L., Lasser, R. & Fanger, M.W. My 43, a monoclonal antibody that reacts with human myeloid cells inhibits monocyte IgA binding and triggers function. J. Immunol. 143, 4117–4122 (1989).
Monteiro, R.C., Kubagawa, H. & Cooper, M.D. Cellular distribution, regulation, and biochemical structure of an Fc alpha receptor in humans. J. Exp. Med. 171, 597–613 (1999).
Patry, C., Sibille, Y., Lehuen, A. & Monteiro, R.C. Identification of Fcα receptor (CD89) isoforms generated by alternative splicing that are differently expressed between blood monocytes and alveolar macrophages. J. Immunol. 156, 4442–4448 (1996).
Nikolova, E.B., Tomana, M. & Russell, M.W. The role of the carbohydrate chains in complement (C3) fixation by solid-phase-bound human IgA. Immunology 82, 321–327 (1994).
van Egmond, M. et al. IgA and the IgA Fc receptor. Trends Immunol. 22, 205–211 (2001).
Dalia, A.B. & Weiser, J.N. Minimization of bacterial size allows for complement evasion and is overcome by the agglutinating effect of antibody. Cell Host Microbe 10, 486–496 (2011).
Thiesen, B., Greenwood, B., Brieske, N. & Achtman, M. Persistence of antibodies to meningococcal IgA1 protease versus decay of antibodies to group A polysaccharide and Opc protein. Vaccine 15, 209–219 (1997).
Kobayashi, K., Fujiyama, Y., Hagiwara, K. & Kondoh, H. Resistance of normal serum IgA and secretory IgA to bacterial IgA proteases: evidence for the presence of enzyme-neutralizing antibodies in both serum and secretory IgA, and also in serum IgG. Microbiol. Immunol. 31, 1097–1106 (1987).
Plaut, A.G., Qiu, J., Grundy, F. & Wright, A. Growth of Haemophilus influenzae in human milk: synthesis, distribution, and activity of IgA protease as determined by study of iga+ and mutant iga- cells. J. Infect. Dis. 166, 43–52 (1992).
Devenyi, A.G., Plaut, A.G., Grundy, F.J. & Wright, A. Post-infectious human serum antibodies inhibit IgA1 proteinases by interaction with the cleavage site specificity determinant. Mol. Immunol. 30, 1243–1248 (1993).
Romanello, V. et al. Cloning, expression, purification, and characterization of Streptococcus pneumoniae IgA1 protease. Protein Expr. Purif. 45, 142–149 (2006).
Reinholdt, J. & Kilian, M. Comparative analysis of immunoglobulin A1 protease activity among bacteria representing different genera, species, and strains. Infect. Immun. 65, 4452–4459 (1997).
Musher, D.M. et al. Emergence of antibody to capsular polysaccharides of Streptococcus pneumoniae during outbreaks of pneumonia: association with nasopharyngeal colonization. Clin. Infect. Dis. 24, 441–446 (1997).
Blake, M.S. & Swanson, J. Studies on gonococcus infection. XVI. Purification of Neisseria gonorrhoeae immunoglobulin A1 protease. Infect. Immun. 22, 350–358 (1978).
Carroll, W.L., Thielemans, K., Dilley, J. & Levy, R. Mouse × human heterohybridomas as fusion partners with human B cell tumors. J. Immunol. Methods 89, 61–72 (1986).
Lefkovits, I. & Waldmann, H. Limiting dilution analysis of the cell of immune system I. The clonal basis of immune response. Immunol. Today 5, 265–268 (1984).
Johnson, S., Opstad, N.L., Douglas, J.M. Jr. & Janoff, E.N. Prolonged and preferential production of polymeric immunoglobulin A in response to Streptococcus pneumoniae capsular polysaccharides. Infect. Immun. 64, 4339–4344 (1996).
Rubins, J.B. et al. Dual function of pneumolysin in the early pathogenesis of murine pneumococcal pneumonia. J. Clin. Invest. 95, 142–150 (1995).
Acknowledgements
All human and mouse studies were approved by the Human and Animal Subjects Subcommittees, and written informed consent was obtained from patients. We thank Ann Emery for secretarial support. This work was supported by the National Institutes of Health Grants AI-092468, AI-48796, AI-42240, HD-41361, AI092468 (ENJ), AI-38446 (JNW), DE15844, the Tufts Digestive Disease Research Center P30 DK34928 (AGP), the Mucosal and Vaccine Research Center at the University of Colorado School of Medicine, and the Department of Veterans Affairs Research Service.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Additional information
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Supplementary information
Rights and permissions
About this article
Cite this article
Janoff, E., Rubins, J., Fasching, C. et al. Pneumococcal IgA1 protease subverts specific protection by human IgA1. Mucosal Immunol 7, 249–256 (2014). https://doi.org/10.1038/mi.2013.41
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2013.41
This article is cited by
-
Pneumococcal capsule blocks protection by immunization with conserved surface proteins
npj Vaccines (2021)
-
Similar genomic patterns of clinical infective endocarditis and oral isolates of Streptococcus sanguinis and Streptococcus gordonii
Scientific Reports (2020)
-
Within-host microevolution of Streptococcus pneumoniae is rapid and adaptive during natural colonisation
Nature Communications (2020)
-
Topical application of nebulized human IgG, IgA and IgAM in the lungs of rats and non-human primates
Respiratory Research (2019)
-
Characterization of the zinc metalloprotease of Streptococcus suis serotype 2
Veterinary Research (2018)