Abstract
Nucleic acid-based (molecular) analytical methods have utility in discriminating between bowel microbiota of altered compositions. This has been demonstrated in studies involving both experimental animals and humans with inflammation of the bowel. Although alterations in the composition of the microbiota can be demonstrated, future studies need to provide functional links between the candidate proinflammatory agents and disease processes.
Similar content being viewed by others
Introduction
Numerous microbial populations, mostly bacterial, that interact to form a community of considerable biomass, biodiversity, and stability, inhabit the large bowels of humans. This community, often referred to as the microflora or microbiota, digests complex polymers derived from the host's food (such as dietary fiber) and from the host's secretions (such as mucins in mucus). Fermentation of the molecules resulting from the hydrolysis of polysaccharides and proteins produces short chain fatty acids, gases, phenols, indoles, and amines as major products.1 As the sustenance of the bacteria is dependent on the host, the bowel can be likened to a banqueting table that the human host shares with its bacterial guests. Thus the resident bacteria are also referred to as “commensals”. The fermentation products, plus the incalculable antigenic load associated with the bacterial cells, affect the development and maintenance of physiological processes.2, 3, 4 and 5 The bowel community, whole or in part, may act as a surrogate pathogen in inflammatory bowel diseases (IBD). Continuous challenge of the mucosal immune system by bacterial antigens as a result of abnormal epithelial permeability may drive the chronic immune inflammation observed in IBD.6
Results
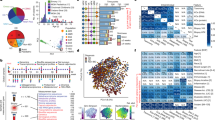
Examples of the outcome of PCR/DGGE and fluorescence in situ hybridization (FISH) analyses of bowel samples are shown in Figure 1. Evidence of the utility of these methods for analysis of the composition of the microbiota is provided by the following examples.
Examples of results of analytical methods. (a and b) Microbiota profiles generated by PCR/DGGE. Numbers indicate different patients from whom biopsies of inflamed (In) and noninflamed (N) bowel tissues were obtained. (c–h) Fluorescence in situ hybridization (FISH)/flow cytometry analysis of human stool. (c) R1/M1 region of histogram, bacterial cells reacted with universal bacterial probe. (d) M2, absence of fluorescent bacterial cells when exposed to negative control probe. (e) Absence of fluorescent bacterial cells in scatter plot in R2 region (negative control). (g) M4 region of histogram contains bacterial cells reacted with universal probe and group-specific probe. Counts of double-labeled cells can be expressed as percentage of those obtained with universal probe alone (f). (h) Scatter plot showing bacterial cells in region R4 double-labeled with universal and group-specific probes.
The bacteriology of biopsies differs between newly diagnosed, untreated Crohn's disease and ulcerative colitis patients
Biopsies were collected from inflamed and noninflamed sites in the intestines of newly diagnosed, untreated patients. Biopsies were also collected from several intestinal sites of healthy subjects. The bacterial collections associated with the biopsies were analyzed by generating PCR/DGGE profiles, the preparation of 16S rRNA gene clone libraries, and qualitative PCR to detect specific groups of bacteria. The total numbers of bacteria associated with the biopsies were determined by real-time quantitative PCR. Ulcerative colitis patients had more bacteria associated with biopsies than the Crohn's disease patients and healthy subjects. The composition of the bacterial collections in ulcerative colitis and Crohn's disease patients differed. Unclassified members of the phyla Bacteroidetes and Verrucomicrobia were more prevalent in Crohn's disease than in ulcerative colitis patients. The profiles did not differ between inflamed and noninflamed tissues.7
Differential clustering of bowel biopsy-associated bacterial profiles of specimens collected in Mexico and Canada
Bacterial collections associated with bowel biopsies, aspirates of residual fluid after bowel cleansing, and feces from IBD patients and healthy subjects in Edmonton, Canada and Mexico City, Mexico were investigated. PCR/TTGE analysis of 16S rRNA gene sequences produced profiles of the bacterial collections whose similarities were compared. Similarity analysis showed that the profiles did not cluster according to disease status, but that Canadian and Mexican profiles could be differentiated by this method. Comparison of biopsy, aspirate, and fecal samples obtained from the same subject showed that, on average, the profiles were highly similar. Therefore biopsy-associated bacteria are likely to represent, at least in part, contaminants from the fluid, which resembles a fecal solution, that pools in the bowel after cleansing before endoscopy.8
Bacteriological content of pelvic pouches of pouchitis and familial adenomatous polyposis patients, and impact of antibiotic therapy
Chronic or recurrent pouchitis is the most important long-term complication leading to poor function following ileal pouch-anal anastomosis for ulcerative colitis. Antibiotic administration reduces symptoms of pouchitis indicating that bacteria have a role in pathogenesis. The stool microbiota of patients with pouchitis and of familial adenomatous polyposis patients were analyzed by PCR/TTGE and FISH combined with flow cytometry. The composition of the stool of pouchitis and familial adenomatous polyposis patients was markedly different. Electrophoretic profiles of the stool microbiota of familial adenomatous patients clustered at the 80% level of similarity, whereas those of pouchitis patients were disparate. The results of FISH analysis showed that bacteria not commonly present in human feces, nor in the stool of familial adenomatous polyposis patients, comprised about 50% of the stool microbiota of untreated pouchitis patients. Antibiotic treatment reduced the proportion of these unknown bacteria in the stool of pouchitis patients. Therefore chronic or recurrent pouchitis is associated with a microbiota that contains bacteria not commonly associated with human feces or FAP pouches. These bacteria have a reduced prevalence when antibiotics are administered and therefore are likely to be the pathogenic agents of pouchitis (Tannock G.W. et al., unpublished data).
Analysis of the large bowel microbiota of colitic mice using PCR/DGGE
The colonic microbiota of formerly germfree interleukin 10 (IL-10)-deficient mice that had been exposed to the fecal microbiota of specific-pathogen-free animals was screened using PCR/DGGE. The composition of the large bowel microbiota of IL-10-deficient mice changed as colitis progressed. DNA fragments originating from four bacterial populations (“Bacteroides sp.”, Bifidobacterium animalis, Clostridium cocleatum, enterococci) were more apparent in PCR/DGGE profiles of colitic mice relative to noncolitic animals, whereas two populations were less apparent (Eubacterium ventriosum, Acidophilus group lactobacilli). Specific DNA/RNA dot blot analysis showed that bifidobacterial rRNA abundance increased as colitis developed. PCR/DGGE was therefore shown to be an effective method to demonstrate changes in the composition of the large bowel microbiota of mice in relation to progression of inflammatory disease.9 Monoassociation of IL-10-deficient mice with B. animalis subspecies animalis resulted in bowel inflammation, completing Koch's postulates.10
Discussion
The results of nucleic acid-based analyses provide targets for further investigations in which functional links need to be established between the candidate proinflammatory bacteria and disease. Culture of the not-yet-cultivable bacteria resident in the human bowel must therefore have priority, otherwise alterations in microbiota composition will remain merely statistical correlations and cause-and-effect will never be established.
Methods
Nucleic acid-based (molecular) analytical methods have been used to assess the composition of the bacterial community because most of its members have not yet been cultivated in the laboratory using traditional bacteriological methods. Although the molecular methods have assisted in defining and comparing bacterial communities in general terms, the phylogenetic information is often shallow (broad phylogenetic groups) and outcomes of investigations are confounded by interindividual and international differences in the composition of bowel communities, a polluted database in which about 5% of entries are reported to be invalid, and biases inherent in sampling (subjects and specimens) as well as in analytical procedures.11, 12, 13 and 14
Nevertheless, three analytical methods show utility in investigations of the bowel microbiota in relation to IBD: phylogenetic catalogs of small ribosomal subunit RNA (16S rRNA in bacteria) gene sequences that delineate molecular species, detection and enumeration of bacterial groups using fluorescent oligonucleotide probes that target ribosomal RNA sequences, and the preparation of microbiota “snapshots” by means of a combination of PCR and denaturing polyacrylamide gel electrophoresis.
The starting point for nucleic acid-based methods is the extraction of bacterial DNA or RNA directly from the fecal or other sample of interest, avoiding the need to cultivate any members of the bacterial community. 16S rRNA or the gene that encodes it has become a cornerstone of bacterial classification because it contains regions of nucleotide base sequence that are highly conserved across the bacterial world and that are interspersed with variable regions (V regions). These V regions contain the “signatures” of phylogenetic groups and even species.15 For this reason, V regions of 16S rRNA (or 16S rRNA gene sequences) are the basis of the molecular analytical methods. Bacterial DNA or RNA extracted from the samples (in theory, nucleic acid from all of the bacterial types in the sample will be represented in the extracts) and PCR amplification (reverse transcription–PCR in the case of RNA extracts) of the 16S rRNA gene, in part or complete, is carried out. Clone libraries of the 16S rRNA genes can be made and sequenced, or emulsion-based high-throughput PCR sequencing can be carried out, producing a catalog of the bacterial constituents of the ecosystem.16, 17
From the information in databases, DNA (oligonucleotide) probes can be designed and synthesized. They specifically target V regions of the 16S or 23S rRNA molecules within ribosomes.18, 19 and 20 The probes are 5′ labeled with a fluorescent dye which permits both detection and quantification of specific bacterial populations. Bacterial cells within which hybridization with a probe has occurred fluoresce and hence can be detected and counted by epifluorescence microscopy (preferably automated) or fluorescence-activated flow cytometry. Permeabilization of the bacterial cells is required to standardize intracellular access of DNA probes to their targets.21 Important technical considerations include (1) the physiological state of the bacterial cells because the number of ribosomes per bacterial cell is greater the higher the metabolic activity. Therefore, bacterial cells in a quiescent state have weak fluorescence and may not be detected. (2) Another technical consideration is the degree of hybridization stringency, which depends on three factors: temperature, salt concentration, and formamide concentration of the hybridization solution. The manipulation of these factors influences the specificity of hybridization and hence detection and quantification. (3) A third technical consideration is a 16S rRNA gene database that is rapidly increasing is size. More than 600,000 16S rRNA sequences are available from the Ribosomal Database (http://rdp.cme.msu.edu), permitting the in silico development and validation of a large panel of probes targeting different phylotypes resident in the human bowel. Several of the currently used probes were designed and tested using older versions of the Ribosomal Database, so continual reassessment of specificity and coverage of these probes is essential to update and confirm their continuing reliability. (4) The last technical consideration is detection and enumeration. Epifluorescence microscopic detection and quantification of bacterial populations was used originally but, because of the laborious and time consuming nature of this work, automated systems have been developed, culminating in the use of flow cytometry to count the fluorescent cells. Rapid and easy to set up, flow cytometry combines quantitative and multiparametrics analysis (size, internal granularity, fluorescence signal). A lower threshold of detection of 0.4% relative to the total number of bacteria determined with a universal bacterial probe, EUB338, has been reported. This analytical approach is referred to as FISH.
Deriving a catalog of bowel inhabitants, or enumerating groups of bacteria using DNA probes for every sample that needs to be investigated, is a daunting task. A relatively simple, semiquantitative screening method to compare the bacterial composition of multiple samples is provided by PCR combined with denaturing gradient gel electrophoresis. This approach, which provides comparative “snapshots” of microbiota compositions, has been demonstrated to have enormous utility in a number of bowel microbiota studies.7, 22, 23, 24, 25 and 26 DNA or RNA is extracted directly from intestinal or fecal samples. Then a variable 16S rRNA gene sequence is amplified using PCR primers that anneal with conserved sequences that span the selected V region. One of the PCR primers has a GC-rich 5′ end (GC clamp) to prevent complete denaturation of the DNA fragments during gradient gel electrophoresis. To separate the 16S fragments amplified from different types of bacteria and present in the PCR product, a polyacrylamide gel is used. The double-stranded 16S fragments migrate through a polyacrylamide gel containing a chemical (DGGE) or thermal (TTGE) gradient until they are partially denatured by the chemical or temperature conditions. The fragments do not completely denature because of the GC clamp, and migration is radically slowed when partial denaturation occurs. Because of the variation in the 16S sequences of different bacterial species, their chemical and thermal stability is also different; therefore different 16S “species” can be differentiated on the basis of gel migration distances of DNA fragments by this electrophoretic method. Profiles of the numerically predominant members of the microbiota are generated.11, 27 Individual fragments of DNA can be cut from electrophoretic gels, further amplified and cloned, then sequenced. The sequence can be compared to those in gene databanks to obtain identification of the bacterium from which the 16S sequence originated. In a further development of this methodology, PCR primers specific for bacterial groups can be derived. These primers generate an electrophoretic profile of the species comprising a specific bacterial genus, for example bifidobacterial species, within the bacterial community.28, 29 and 30
Disclosure
Gerald W. Tannock has received a grant from the Foundation for Research, Science and Technology.
References
Cummings, J.H. & Macfarlane, G.T. The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70, 443–459 (1991).
Hooper, L.V. et al. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 96, 9833–9838 (1999).
Hooper, L.V. et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884 (2001).
Stappenbeck, T.S. et al. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 99, 15451–15455 (2002).
Backhed, F. et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 101, 15718–15723 (2004).
Macdonald, T.T. & Monteleone, G. Immunity, inflammation, and allergy in the gut. Science 307, 1920–1925 (2005).
Bibiloni, R. et al. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J. Med. Microbiol. 55, 1141–1149 (2006).
Bibiloni, R. et al. Differential clustering of bowel biopsy-associated bacterial profiles of specimens collected in Mexico and Canada: what do these profiles represent. J. Med. Microbiol. 57, 111–117 (2008).
Bibiloni, R. et al. Analysis of the large bowel microbiota of colitic mice using PCR/DGGE. Lett. Appl. Microbiol. 41, 45–51 (2005).
Moran, J.P. et al. Bifidobacterium animalis causes extensive duodenitis and mild colonic inflammation in interleukin-10 knockout mice. Gastroenterology 130, A6 (2006).
Zoetendal, E.G. et al. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host specific communities of active bacteria. Appl. Environ. Microbiol. 64, 3854–3859 (1998).
Ashelford, K.E. et al. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71, 7724–7736 (2005).
Eckburg, P.B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Ashelford, K.E. et al. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72, 5734–57341 (2006).
Woese, C.R. Bacterial evolution. Microbiol. Rev. 51, 221–271 (1987).
Suau, A. et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65, 4799–4807 (1999).
Sogin, M.L. et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA. 103, 12115–12120 (2006).
Franks, A. et al. Variations of bacterial population in human faeces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64, 3336–3345 (1998).
Sghir, A. et al. Quantification of bacterial groups within the human faecal flora by oligonucleotide probe hybridisation. Appl. Environ. Microbiol. 66, 2263–2266 (2000).
Lay, C. et al. Colonic microbiota signatures across five northern European countries. Appl. Environ. Microbiol. 71, 4153–4155 (2005).
Lay, C. et al. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ. Microbiol. 7, 933–946 (2005).
Guan, L.L. et al. Detection and identification of Lactobacillus species in crops of broilers of different ages by using PCR-denaturing gradient gel electrophoresis and amplified ribosomal DNA restriction analysis. Appl. Environ. Microbiol. 69, 6750–7 (2003).
Schultz, M. et al. Effects of feeding a probiotic preparation (SIM) containing inulin on the severity of colitis and on the composition of the intestinal microflora in HLA-B27 transgenic rats. Clin. Diag. Lab. Immunol. 11, 581–587 (2004).
Tannock, G.W. et al. Impact of consumption of oligosaccharide-containing biscuits on the fecal microbiota of humans. Appl. Environ. Microbiol. 70, 2129–2136 (2004).
Hoentjen, F. et al. Reduction of colitis by prebiotics in HLA-B27 transgenic rats is associated with microflora changes and immunomodulation. Inflamm. Bowel Dis. 11, 977–985 (2005).
Snart, J. et al. Supplementation of the diet with high-viscosity beta-glucan results in enrichment for lactobacilli in the rat cecum. Appl. Environ. Microbiol. 72, 1925–1931 (2006).
Muyzer, G. & Smalla, K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leewenhoek 73, 127–141 (1998).
Walter, J. et al. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human faeces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67, 2578–2585 (2001).
Knarreborg, A. et al. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 68, 5918–24 (2002).
Requena, T. et al. Identification, detection, and enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 68, 2420–2427 (2002).
Acknowledgements
We gratefully acknowledge the support of the Crohn's and Colitis Foundations of America and of Canada, the Lottery Grants Board, the HealthCare Otago Charitable Trust, the Otago Medical Research Foundation, the Otago University Research Committee, and the Otago Schools of Medical Science and of Medicine to the research described in this article.
Author information
Authors and Affiliations
Corresponding author
PowerPoint slides
Rights and permissions
About this article
Cite this article
Tannock, G. Molecular analysis of the intestinal microflora in IBD. Mucosal Immunol 1 (Suppl 1), S15–S18 (2008). https://doi.org/10.1038/mi.2008.54
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2008.54
This article is cited by
-
Effect of the standard herbal preparation, STW5, treatment on dysbiosis induced by dextran sodium sulfate in experimental colitis
BMC Complementary Medicine and Therapies (2021)
-
Development of gut inflammation in mice colonized with mucosa-associated bacteria from patients with ulcerative colitis
Gut Pathogens (2015)
-
Mucosal immunity in human and simian immunodeficiency lentivirus infections
Mucosal Immunology (2013)
-
Impact of Intestinal Microbiota on Intestinal Luminal Metabolome
Scientific Reports (2012)
-
Probiotics and the gut microbiota in intestinal health and disease
Nature Reviews Gastroenterology & Hepatology (2010)