Abstract
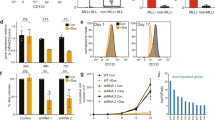
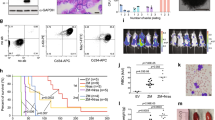
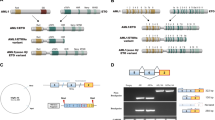
Overexpression of the BRE (brain and reproductive organ-expressed) gene defines a distinct pediatric and adult acute myeloid leukemia (AML) subgroup. Here we identify a promoter enriched for active chromatin marks in BRE intron 4 causing strong biallelic expression of a previously unknown C-terminal BRE transcript. This transcript starts with BRE intron 4 sequences spliced to exon 5 and downstream sequences, and if translated might code for an N terminally truncated BRE protein. Remarkably, the new BRE transcript was highly expressed in over 50% of 11q23/KMT2A (lysine methyl transferase 2A)-rearranged and t(8;16)/KAT6A-CREBBP cases, while it was virtually absent from other AML subsets and normal tissues. In gene reporter assays, the leukemia-specific fusion protein KMT2A-MLLT3 transactivated the intragenic BRE promoter. Further epigenome analyses revealed 97 additional intragenic promoter marks frequently bound by KMT2A in AML with C-terminal BRE expression. The corresponding genes may be part of a context-dependent KMT2A-MLLT3-driven oncogenic program, because they were higher expressed in this AML subtype compared with other groups. C-terminal BRE might be an important contributor to this program because in a case with relapsed AML, we observed an ins(11;2) fusing CHORDC1 to BRE at the region where intragenic transcription starts in KMT2A-rearranged and KAT6A-CREBBP AML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hinai AA, Valk PJ . Review: aberrant EVI1 expression in acute myeloid leukaemia. Br J Haematol 2016; 172: 870–878.
Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 2014; 157: 369–381.
Yamazaki H, Suzuki M, Otsuki A, Shimizu R, Bresnick EH, Engel JD et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell 2014; 25: 415–427.
Senyuk V, Sinha KK, Li D, Rinaldi CR, Yanamandra S, Nucifora G . Repression of RUNX1 activity by EVI1: a new role of EVI1 in leukemogenesis. Cancer Res 2007; 67: 5658–5666.
Laricchia-Robbio L, Fazzina R, Li D, Rinaldi CR, Sinha KK, Chakraborty S et al. Point mutations in two EVI1 Zn fingers abolish EVI1-GATA1 interaction and allow erythroid differentiation of murine bone marrow cells. Mol Cell Biol 2006; 26: 7658–7666.
Laricchia-Robbio L, Premanand K, Rinaldi CR, Nucifora G . EVI1 Impairs myelopoiesis by deregulation of PU.1 function. Cancer Res 2009; 69: 1633–1642.
Swansbury GJ, Slater R, Bain BJ, Moorman AV, Secker-Walker LM . Hematological malignancies with t(9;11)(p21-22;q23)—a laboratory and clinical study of 125 cases. European 11q23 Workshop participants. Leukemia 1998; 12: 792–800.
Rubnitz JE, Raimondi SC, Tong X, Srivastava DK, Razzouk BI, Shurtleff SA et al. Favorable impact of the t(9;11) in childhood acute myeloid leukemia. J Clin Oncol 2002; 20: 2302–2309.
Arai S, Yoshimi A, Shimabe M, Ichikawa M, Nakagawa M, Imai Y et al. Evi-1 is a transcriptional target of mixed-lineage leukemia oncoproteins in hematopoietic stem cells. Blood 2011; 117: 6304–6314.
Noordermeer SM, Monteferrario D, Sanders MA, Bullinger L, Jansen JH, van der Reijden BA . Improved classification of MLL-AF9-positive acute myeloid leukemia patients based on BRE and EVI1 expression. Blood 2012; 119: 4335–4337.
Balgobind BV, Zwaan CM, Reinhardt D, Arentsen-Peters TJ, Hollink IH, de Haas V et al. High BRE expression in pediatric MLL-rearranged AML is associated with favorable outcome. Leukemia 2010; 24: 2048–2055.
Noordermeer SM, Sanders MA, Gilissen C, Tonnissen E, van der Heijden A, Dohner K et al. High BRE expression predicts favorable outcome in adult acute myeloid leukemia, in particular among MLL-AF9-positive patients. Blood 2011; 118: 5613–5621.
Chui YL, Ma CH, Li W, Xu Z, Yao Y, Lin FK et al. Anti-apoptotic protein BRE/BRCC45 attenuates apoptosis through maintaining the expression of caspase inhibitor XIAP in mouse Lewis lung carcinoma D122 cells. Apoptosis 2014; 19: 829–840.
Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell 2003; 12: 1087–1099.
Chan JY, Li L, Miao J, Cai DQ, Lee KK, Chui YL . Differential expression of a novel gene BRE (TNFRSF1A modulator/BRCC45) in response to stress and biological signals. Mol Biol Rep 2010; 37: 363–368.
Shi W, Tang MK, Yao Y, Tang C, Chui YL, Lee KK . BRE plays an essential role in preventing replicative and DNA damage-induced premature senescence. Scientific Rep 2016; 6: 23506.
Feng L, Huang J, Chen J . MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev 2009; 23: 719–728.
Patterson-Fortin J, Shao G, Bretscher H, Messick TE, Greenberg RA . Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J Biol Chem 2010; 285: 30971–30981.
Kyrieleis OJ, McIntosh PB, Webb SR, Calder LJ, Lloyd J, Patel NA et al. Three-dimensional architecture of the human BRCA1-A histone deubiquitinase core complex. Cell Rep 2016; 17: 3099–3106.
Yan K, Li L, Wang X, Hong R, Zhang Y, Yang H et al. The deubiquitinating enzyme complex BRISC is required for proper mitotic spindle assembly in mammalian cells. J Cell Biol 2015; 210: 209–224.
Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004; 350: 1617–1628.
Haferlach T, Kohlmann A, Klein HU, Ruckert C, Dugas M, Williams PM et al. AML with translocation t(8;16)(p11;p13) demonstrates unique cytomorphological, cytogenetic, molecular and prognostic features. Leukemia 2009; 23: 934–943.
Coenen EA, Zwaan CM, Reinhardt D, Harrison CJ, Haas OA, de Haas V et al. Pediatric acute myeloid leukemia with t(8;16)(p11;p13), a distinct clinical and biological entity: a collaborative study by the International-Berlin-Frankfurt-Munster AML-study group. Blood 2013; 122: 2704–2713.
Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R . Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009; 113: 3088–3091.
Prange KH, Mandoli A, Kuznetsova T, Wang SY, Sotoca AM, Marneth AE et al. MLL-AF9 and MLL-AF4 oncofusion proteins bind a distinct enhancer repertoire and target the RUNX1 program in 11q23 acute myeloid leukemia. Oncogene 2017; 36: 3346–3356.
Di Savino A, Panuzzo C, Rocca S, Familiari U, Piazza R, Crivellaro S et al. Morgana acts as an oncosuppressor in chronic myeloid leukemia. Blood 2015; 125: 2245–2253.
Castilla LH, Perrat P, Martinez NJ, Landrette SF, Keys R, Oikemus S et al. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proc Natl Acad Sci USA 2004; 101: 4924–4929.
Landrette SF, Kuo YH, Hensen K, Barjesteh van Waalwijk van Doorn-Khosrovani S, Perrat PN, Van de Ven WJ et al. Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb-MYH11. Blood 2005; 105: 2900–2907.
Slany RK . The molecular mechanics of mixed lineage leukemia. Oncogene 2016; 35: 5215–5223.
Marschalek R . Systematic classification of mixed-lineage leukemia fusion partners predicts additional cancer pathways. Ann Lab Med 2016; 36: 85–100.
Chan EM, Chan RJ, Comer EM, Goulet RJ, Crean CD, Brown ZD et al. MOZ and MOZ-CBP cooperate with NF-kappaB to activate transcription from NF-kappaB-dependent promoters. Exp Hematol 2007; 35: 1782–1792.
Paggetti J, Largeot A, Aucagne R, Jacquel A, Lagrange B, Yang XJ et al. Crosstalk between leukemia-associated proteins MOZ and MLL regulates HOX gene expression in human cord blood CD34+ cells. Oncogene 2010; 29: 5019–5031.
Katsumoto T, Yoshida N, Kitabayashi I . Roles of the histone acetyltransferase monocytic leukemia zinc finger protein in normal and malignant hematopoiesis. Cancer Sci 2008; 99: 1523–1527.
Kitabayashi I, Aikawa Y, Nguyen LA, Yokoyama A, Ohki M . Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J 2001; 20: 7184–7196.
Roy R, Chun J, Powell SN . BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 2011; 12: 68–78.
Zhang J, Cao M, Dong J, Li C, Xu W, Zhan Y et al. ABRO1 suppresses tumourigenesis and regulates the DNA damage response by stabilizing p53. Nat Commun 2014; 5: 5059.
Huang D, Nagata Y, Grossmann V, Radivoyevitch T, Okuno Y, Nagae G et al. BRCC3 mutations in myeloid neoplasms. Haematologica 2015; 100: 1051–1057.
Chan BC, Ching AK, To KF, Leung JC, Chen S LiQ et al. BRE is an antiapoptotic protein in vivo and overexpressed in human hepatocellular carcinoma. Oncogene 2008; 27: 1208–1217.
Chen HB, Pan K, Tang MK, Chui YL, Chen L, Su ZJ et al. Comparative proteomic analysis reveals differentially expressed proteins regulated by a potential tumor promoter, BRE, in human esophageal carcinoma cells. Biochem Cell Biol 2008; 86: 302–311.
Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA et al. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell 2016; 29: 723–736.
Noordermeer SM, Wennemers M, Bergevoet SM, van der Heijden A, Tonnissen E, Sweep FC et al. Expression of the BRCA1 complex member BRE predicts disease free survival in breast cancer. Breast Cancer Res Treat 2012; 135: 125–133.
Acknowledgements
We thank Dr Robert Slany for kindly sharing the pGL3-Hoxa7 promoter plasmid. This work was supported by the Radboud University Medical Center for Oncology, the Dutch Cancer Foundation (KUN 2011-4937) and the European Union’s Seventh Framework Programme (FP5/2007-2013, 282510-BLUEPRINT).
Author contributions
AEM, KHMP, BAR and JHAM coordinated research and wrote the manuscript. AEM, KHMP and SMB conducted and analyzed most experiments. KHMP, EMJM, BK, NS, MLY, JK, HGS and JHAM contributed to ChIP-seq and RNA-sequencing of KMT2A-MLLT3 samples (BLUEPRINT consortium). KHMP, NT and MAS did bioinformatic analyses. ASAAH, ECGS, JK, MAS and PJMV detected CHORDC1-BRE fusion. TCJMA-P, CMZ, HGS, MMH-E, TH, MF, JHJ, PJMV, BAR and JHM provided patient samples and funding. All authors critically revised the paper and approved the final version.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Marneth, A., Prange, K., Al Hinai, A. et al. C-terminal BRE overexpression in 11q23-rearranged and t(8;16) acute myeloid leukemia is caused by intragenic transcription initiation. Leukemia 32, 828–836 (2018). https://doi.org/10.1038/leu.2017.280
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.280
This article is cited by
-
High expression of an intragenic long noncoding RNA misinterpreted as high FTO oncogene expression in NPM1 mutant acute myeloid leukemia
Leukemia (2023)
-
Co-inhibition of HDAC and MLL-menin interaction targets MLL-rearranged acute myeloid leukemia cells via disruption of DNA damage checkpoint and DNA repair
Clinical Epigenetics (2019)
-
Acute myeloid leukemia with t(8;16)(p11.2;p13.3)/KAT6A-CREBBP in adults
Annals of Hematology (2019)