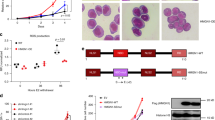
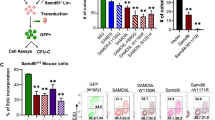
Abstract
The role that changes in DNA methylation and histone modifications have in human malignancies is poorly understood. p300 and CREB-binding protein (CBP), two distinct but highly homologous lysine acetyltransferases, are mutated in several cancers, suggesting their role as tumor suppressors. In the current study, we found that deletion of p300, but not CBP, markedly accelerated the leukemogenesis ofNup98-HoxD13 (NHD13) transgenic mice, an animal model that phenotypically copies human myelodysplastic syndrome (MDS). p300 deletion restored the ability of NHD13 expressing hematopoietic stem and progenitor cells (HSPCs) to self-renew in vitro, and to expand in vivo, with an increase in stem cell symmetric self-renewal divisions and a decrease in apoptosis. Furthermore, loss of p300, but not CBP, promoted cytokine signaling, including enhanced activation of the MAPK and JAK/STAT pathways in the HSPC compartment. Altogether, our data indicate that p300 has a pivotal role in blocking the transformation of MDS to acute myeloid leukemia, a role distinct from that of CBP.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nimer SD . Myelodysplastic syndromes. Blood 2008; 111: 4841–4851.
Maegawa S, Gough SM, Watanabe-Okochi N, Lu Y, Zhang NX, Castoro RJ et al. Age-related epigenetic drift in the pathogenesis of MDS and AML. Genome Res 2014; 24: 580–591.
Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 2011; 364: 2496–2506.
Goodman RH, Smolik S . CBP/p300 in cell growth, transformation, and development. Genes Dev 2000; 14: 1553–1577.
Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, Niedrist D, Papadia F et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet 2005; 76: 572–580.
Iyer NG, Ozdag H, Caldas C . p300/CBP and cancer. Oncogene 2004; 23: 4225–4231.
Borrow J, Stanton Jr VP, Andresen JM, Becher R, Behm FG, Chaganti RS et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet 1996; 14: 33–41.
Kitabayashi I, Aikawa Y, Yokoyama A, Hosoda F, Nagai M, Kakazu N et al. Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia 2001; 15: 89–94.
Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 1998; 93: 361–372.
Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, Livingston DM . Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc Natl Acad Sci USA 2002; 99: 14789–14794.
Chan WI, Hannah RL, Dawson MA, Pridans C, Foster D, Joshi A et al. The Transcriptional coactivator Cbp regulates self-renewal and differentiation in adult hematopoietic stem cells. Mol Cell Biol 2011; 31: 5046–5060.
Kung AL, Rebel VI, Bronson RT, Ch'ng LE, Sieff CA, Livingston DM et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev 2000; 14: 272–277.
Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 2011; 333: 765–769.
Shigeno K, Yoshida H, Pan L, Luo JM, Fujisawa S, Naito K et al. Disease-related potential of mutations in transcriptional cofactors CREB-binding protein and p300 in leukemias. Cancer Lett 2004; 213: 11–20.
Ramos F, Robledo C, Izquierdo-Garcia FM, Suarez-Vilela D, Benito R, Fuertes M et al. Bone marrow fibrosis in myelodysplastic syndromes: a prospective evaluation including mutational analysis. Oncotarget 2016; 7: 30492–30503.
Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013; 122: 3616–3627,quiz 3699.
Duhoux FP, De Wilde S, Ameye G, Bahloula K, Medves S, Lege G et al. Novel variant form of t(11;22)(q23;q13)/MLL-EP300 fusion transcript in the evolution of an acute myeloid leukemia with myelodysplasia-related changes. Leuk Res 2011; 35: e18–e20.
Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong CL, de Pauw A et al. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol 2006; 26: 789–809.
Xu HM, Menendez S, Schlegelberger B, Bae N, Aplan PD, Gohring G et al. Loss of p53 accelerates the complications of myelodysplastic syndrome in a NUP98-HOXD13-driven mouse model. Blood 2012; 120: 3089–3097.
Kanehisa M, Goto S . KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000; 28: 27–30.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Lin YW, Slape C, Zhang Z, Aplan PD . NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood 2005; 106: 287–295.
Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet 2012; 44: 53–U77.
Pan J, Zhang Q, Wang YA, You M . 26S Proteasome activity is down-regulated in lung cancer stem-like cells propagated in vitro. Plos One 2010; 5: e13298.
Offman J, Opelz G, Doehler B, Cummins D, Halil O, Banner NR et al. Defective DNA mismatch repair in acute myeloid leukemia/myelodysplastic syndrome after organ transplantation. Blood 2004; 104: 822–828.
Schinke C, Giricz O, Li WJ, Shastri A, Gordon S, Barreryo L et al. IL8-CXCR2 pathway inhibition as a therapeutic strategy against MDS and AML stem cells. Blood 2015; 125: 3144–3152.
Testa U, Riccioni R, Diverio D, Rossini A, Lo Coco F, Peschle C . Interleukin-3 receptor in acute leukemia. Leukemia 2004; 18: 219–226.
Gits J, van Leeuwen D, Carroll HP, Touw IP, Ward AC . Multiple pathways contribute to the hyperproliferative responses from truncated granulocyte colony-stimulating factor receptors. Leukemia 2006; 20: 2111–2118.
Jordan CT, Upchurch D, Szilvassy SJ, Guzman ML, Howard DS, Pettigrew AL et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia 2000; 14: 1777–1784.
O'Sullivan LA, Liongue C, Lewis RS, Stephenson SEM, Ward AC . Cytokine receptor signaling through the Jak-Stat-Socs pathway in disease. Mol Immunol 2007; 44: 2497–2506.
Vainchenker W, Constantinescu SN . JAK/STAT signaling in hematological malignancies. Oncogene 2013; 32: 2601–2613.
Chung YJ, Park BB, Kang YJ, Kim TM, Eaves CJ, Oh IH . Unique effects of Stat3 on the early phase of hematopoietic stem cell regeneration. Blood 2006; 108: 1208–1215.
Wang L, Gural A, Sun XJ, Zhao XY, Perna F, Huang G et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 2011; 333: 765–769.
Wu M, Kwon HY, Rattis F, Blum J, Zhao C, Ashkenazi R et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell 2007; 1: 541–554.
Van Etten RA . Aberrant cytokine signaling in leukemia. Oncogene 2007; 26: 6738–6749.
Baker SJ, Rane SG, Reddy EP . Hematopoietic cytokine receptor signaling. Oncogene 2007; 26: 6724–6737.
Giotopoulos G, Chan WI, Horton SJ, Ruau D, Gallipoli P, Fowler A et al. The epigenetic regulators CBP and p300 facilitate leukemogenesis and represent therapeutic targets in acute myeloid leukemia. Oncogene 2015; 35: 279–289.
Acknowledgements
We thank the members of the Nimer lab for their assistance and thoughtful input. We thank the Sylvester Comprehensive Cancer Center Flow Cytometry and Oncogenomics Core Facilities helping carry out this work. We also thank Delphine Prou for her assistance in preparing this manuscript. This work was supported by National Cancer Institute Grant R01 CA166835-01 (SN). Shiekhattar’s laboratory was supported by R01 GM078455 and R01 GM105754 from the National Institute of Health and Sylvester Comprehensive Cancer Center.
Author contributions
SDN supervised the project. GC designed and performed most of the experiments, analyzed the data and wrote the manuscript; F Liu analyzed the data and revised the manuscript; TA, MN and F Lai performed some of the experiments and analyzed the data; HX generated the mouse colonies. SC, SG, PJH, KA, CM, MT and LW helped with some of the experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Cheng, G., Liu, F., Asai, T. et al. Loss of p300 accelerates MDS-associated leukemogenesis. Leukemia 31, 1382–1390 (2017). https://doi.org/10.1038/leu.2016.347
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.347
This article is cited by
-
Transcriptional co-activators: emerging roles in signaling pathways and potential therapeutic targets for diseases
Signal Transduction and Targeted Therapy (2023)
-
Molecular characterization of the histone acetyltransferase CREBBP/EP300 genes in myeloid neoplasia
Leukemia (2022)
-
Inhibition of CBP synergizes with the RNA-dependent mechanisms of Azacitidine by limiting protein synthesis
Nature Communications (2021)
-
Peptidomimetic blockade of MYB in acute myeloid leukemia
Nature Communications (2018)