Abstract
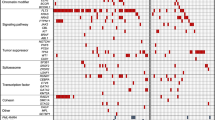
Genomic studies have identified recurrent somatic mutations in acute leukemias. However, current murine models do not sufficiently encompass the genomic complexity of human leukemias. To develop preclinical models, we transplanted 160 samples from patients with acute leukemia (acute myeloid leukemia, mixed lineage leukemia, B-cell acute lymphoblastic leukemia, T-cell ALL) into immunodeficient mice. Of these, 119 engrafted with expected immunophenotype. Targeted sequencing of 374 genes and 265 frequently rearranged RNAs detected recurrent and novel genetic lesions in 48 paired primary tumor (PT) and patient-derived xenotransplant (PDX) samples. Overall, the frequencies of 274 somatic variant alleles correlated between PT and PDX samples, although the data were highly variable for variant alleles present at 0–10%. Seventeen percent of variant alleles were detected in either PT or PDX samples only. Based on variant allele frequency changes, 24 PT-PDX pairs were classified as concordant while the other 24 pairs showed various degree of clonal discordance. There was no correlation of clonal concordance with clinical parameters of diseases. Significantly more bone marrow samples than peripheral blood samples engrafted discordantly. These data demonstrate the utility of developing PDX banks for modeling human leukemia, and emphasize the importance of genomic profiling of PDX and patient samples to ensure concordance before performing mechanistic or therapeutic studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
ADAM. Acute myeloid leukemia. ADAM Medical Encyclopedia 2014 May 29.
Walrath JC, Hawes JJ, Van Dyke T, Reilly KM . Genetically engineered mouse models in cancer research. Adv Cancer Res 2010; 106: 113–164.
Scott CL, Becker MA, Haluska P, Samimi G . Patient-derived xenograft models to improve targeted therapy in epithelial ovarian cancer treatment. Front Oncol 2013; 3: 295.
Malaise M, Neumeier M, Botteron C, Dohner K, Reinhardt D, Schlegelberger B et al. Stable and reproducible engraftment of primary adult and pediatric acute myeloid leukemia in NSG mice. Leukemia 2011; 25: 1635–1639.
Sanchez PV, Perry RL, Sarry JE, Perl AE, Murphy K, Swider CR et al. A robust xenotransplantation model for acute myeloid leukemia. Leukemia 2009; 23: 2109–2117.
Wunderlich M, Mizukawa B, Chou FS, Sexton C, Shrestha M, Saunthararajah Y et al. AML cells are differentially sensitive to chemotherapy treatment in a human xenograft model. Blood 2013; 121: e90–e97.
Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun 2015; 6: 6604.
Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 2011; 469: 362–367.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012; 481: 157–163.
Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012; 150: 264–278.
Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O'Laughlin M et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell 2014; 25: 379–392.
Aparicio S, Hidalgo M, Kung AL . Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer 2015; 15: 311–316.
Vick B, Rothenberg M, Sandhofer N, Carlet M, Finkenzeller C, Krupka C et al. An advanced preclinical mouse model for acute myeloid leukemia using patients' cells of various genetic subgroups and in vivo bioluminescence imaging. PloS One 2015; 10: e0120925.
Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013; 31: 1023–1031.
Cheng DT, Cheng J, Mitchell TN, Syed A, Zehir A, Mensah NY et al. Detection of mutations in myeloid malignancies through paired-sample analysis of microdroplet-PCR deep sequencing data. J Mol Diagn 2014; 16: 504–518.
Li S, Shen D, Shao J, Crowder R, Liu W, Prat A et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep 2013; 4: 1116–1130.
Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015; 43 (Database issue): D805–D811.
He J, Abdel-Wahab O, Nahas MK, Wang K, Rampal RK, Intlekofer AM et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 2016 March, 10.
Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074.
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012; 481: 506–510.
Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010; 464: 999–1005.
Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014; 506: 328–333.
McCormack E, Bruserud O, Gjertsen BT . Review: genetic models of acute myeloid leukaemia. Oncogene 2008; 27: 3765–3779.
Xu D, Liu X, Yu WM, Meyerson HJ, Guo C, Gerson SL et al. Non-lineage/stage-restricted effects of a gain-of-function mutation in tyrosine phosphatase Ptpn11 (Shp2) on malignant transformation of hematopoietic cells. J Exp Med 2011; 208: 1977–1988.
Welcker M, Clurman BE . FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 2008; 8: 83–93.
Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell 2014; 14: 329–341.
Distler E, Wolfel C, Kohler S, Nonn M, Kaus N, Schnurer E et al. Acute myeloid leukemia (AML)-reactive cytotoxic T lymphocyte clones rapidly expanded from CD8(+) CD62L((high)+) T cells of healthy donors prevent AML engraftment in NOD/SCID IL2Rgamma(null) mice. Exp Hematol 2008; 36: 451–463.
Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD Jr et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 2009; 138: 286–299.
Taussig DC, Miraki-Moud F, Anjos-Afonso F, Pearce DJ, Allen K, Ridler C et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood 2008; 112: 568–575.
Mohi MG, Williams IR, Dearolf CR, Chan G, Kutok JL, Cohen S et al. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell 2005; 7: 179–191.
Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 2011; 469: 356–361.
Billerbeck E, Barry WT, Mu K, Dorner M, Rice CM, Ploss A . Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rgamma(null) humanized mice. Blood 2011; 117: 3076–3086.
Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol 2014; 32: 364–372.
Pandey KR, Maden N, Poudel B, Pradhananga S, Sharma AK . The curation of genetic variants: difficulties and possible solutions. Genomics Proteomics Bioinformatics 2012; 10: 317–325.
Stein EM, Altman JK, Collins R, DeAngelo DJ, Fathi AT, Flinn I et al. AG-221, an oral, selective, first-in-class, potent inhibitor of the IDH2 mutant metabolic enzyme, induces durable remissions in a phase I study in patients with IDH2 mutation positive advanced hematologic malignancies. Blood 2014; 2014, ASH abstract (115).
Acknowledgements
We thank Chun-Wei (David) Chen, Omar Abdel-Wahab and Alex Kentsis for critical discussion of the manuscript, Barb Every for editor help. Research funding was provided by the Kleberg Foundation and R01 CA176745/CA/NCI NIH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
KW, MKN, JH, ALD, GMF, DL, SR, PJS, EMS, TB, GAO, RY, VAM are employees and equity holders, RLL is a consultant for Foundation Medicine, Inc.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, K., Sanchez-Martin, M., Wang, X. et al. Patient-derived xenotransplants can recapitulate the genetic driver landscape of acute leukemias. Leukemia 31, 151–158 (2017). https://doi.org/10.1038/leu.2016.166
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.166
This article is cited by
-
Comparison of clonal architecture between primary and immunodeficient mouse-engrafted acute myeloid leukemia cells
Nature Communications (2022)
-
In Vivo Modeling of Human Breast Cancer Using Cell Line and Patient-Derived Xenografts
Journal of Mammary Gland Biology and Neoplasia (2022)
-
Pediatric T-ALL type-1 and type-2 relapses develop along distinct pathways of clonal evolution
Leukemia (2022)
-
PDX models reflect the proteome landscape of pediatric acute lymphoblastic leukemia but divert in select pathways
Journal of Experimental & Clinical Cancer Research (2021)
-
Examining treatment responses of diagnostic marrow in murine xenografts to predict relapse in children with acute lymphoblastic leukaemia
British Journal of Cancer (2020)