Abstract
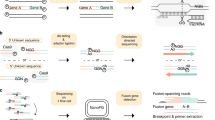
DNA sequence enrichment from complex genomic samples using microarrays enables targeted next-generation sequencing (NGS). In this study, we combined 454 shotgun pyrosequencing with long oligonucleotide sequence capture arrays. We demonstrate the detection of mutations including point mutations, deletions and insertions in a cohort of 22 patients presenting with acute leukemias and myeloid neoplasms. Importantly, this one-step methodological procedure also allowed the detection of balanced chromosomal aberrations, including translocations and inversions. Moreover, the genomic representation of only one of the partner genes of a chimeric fusion on the capture platform also permitted identification of the novel fusion partner genes. Using acute myeloid leukemias harboring RUNX1 abnormalities as a model system, three novel chromosomal fusion sequences and KCNMA1 as a novel RUNX1 fusion partner gene were detected. This assay has the strong potential to become an important method for the comprehensive genetic characterization of particular leukemias and other malignancies harboring complex genomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods 2007; 4: 903–905.
Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 2009; 27: 182–189.
Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol 2009; 27: 1025–1031.
Bacher U, Schnittger S, Haferlach C, Haferlach T . Molecular diagnostics in acute leukemias. Clin Chem Lab Med 2009; 47: 1333–1341.
Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European Leukemia Net. Blood 2010; 115: 453–474.
Pui CH, Evans WE . Treatment of acute lymphoblastic leukemia. N Engl J Med 2006; 354: 166–178.
Grimwade D, Jovanovic JV, Hills RK, Nugent EA, Patel Y, Flora R et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol 2009; 27: 3650–3658.
Ommen HB, Schnittger S, Jovanovic JV, Ommen IB, Hasle H, Ostergaard M et al. Strikingly different molecular relapse kinetics in NPM1c, PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11 acute myeloid leukemias. Blood 2010; 115: 198–205.
Schnittger S, Weisser M, Schoch C, Hiddemann W, Haferlach T, Kern W . New score predicting for prognosis in PML-RARA+, AML1-ETO+, or CBFBMYH11+ acute myeloid leukemia based on quantification of fusion transcripts. Blood 2003; 102: 2746–2755.
Maki K, Yamagata T, Mitani K . Role of the RUNX1-EVI1 fusion gene in leukemogenesis. Cancer Sci 2008; 99: 1878–1883.
Matsuno N, Osato M, Yamashita N, Yanagida M, Nanri T, Fukushima T et al. Dual mutations in the AML1 and FLT3 genes are associated with leukemogenesis in acute myeloblastic leukemia of the M0 subtype. Leukemia 2003; 17: 2492–2499.
De BE, Ferec C, De BM . RUNX1 translocations in malignant hemopathies. Anticancer Res 2009; 29: 1031–1037.
Tang JL, Hou HA, Chen CY, Liu CY, Chou WC, Tseng MH et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood 2009; 114: 5352–5361.
Erben P, Gosenca D, Muller MC, Reinhard J, Score J, Del VF et al. Screening for diverse PDGFRA or PDGFRB fusion genes is facilitated by generic quantitative reverse transcriptase polymerase chain reaction analysis. Haematologica 2010; 95: 738–744.
Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J et al. New insights to the MLL recombinome of acute leukemias. Leukemia 2009; 23: 1490–1499.
Loffler H, Rastetter J . Atlas of Clinical Hematology. Springer, Berlin, 1999.
Schoch C, Schnittger S, Bursch S, Gerstner D, Hochhaus A, Berger U et al. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia 2002; 16: 53–59.
Kern W, Voskova D, Schoch C, Hiddemann W, Schnittger S, Haferlach T . Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood 2004; 104: 3078–3085.
Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 2002; 100: 59–66.
Paulsson K, Horvat A, Strombeck B, Nilsson F, Heldrup J, Behrendtz M et al. Mutations of FLT3, NRAS, KRAS, and PTPN11 are frequent and possibly mutually exclusive in high hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 2008; 47: 26–33.
Bacher U, Haferlach C, Kern W, Haferlach T, Schnittger S . Prognostic relevance of FLT3-TKD mutations in AML: the combination matters—an analysis of 3082 patients. Blood 2008; 111: 2527–2537.
Kohl TM, Schnittger S, Ellwart JW, Hiddemann W, Spiekermann K . KIT exon 8 mutations associated with core-binding factor (CBF)-acute myeloid leukemia (AML) cause hyperactivation of the receptor in response to stem cell factor. Blood 2005; 105: 3319–3321.
Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 2002; 100: 59–66.
Schnittger S, Schoch C, Kern W, Mecucci C, Tschulik C, Martelli MF et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood 2005; 106: 3733–3739.
Schnittger S, Kohl TM, Haferlach T, Kern W, Hiddemann W, Spiekermann K et al. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood 2006; 107: 1791–1799.
Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E et al. Ensembl 2009. Nucleic Acids Res 2009; 37: D690–D697.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437: 376–380.
Li H, Durbin R . Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595.
Shen Y, Wan Z, Coarfa C, Drabek R, Chen L, Ostrowski EA et al. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res 2010; 20: 273–280.
Hasin Y, Olender T, Khen M, Gonzaga-Jauregui C, Kim PM, Urban AE et al. High-resolution copy-number variation map reflects human olfactory receptor diversity and evolution. PLoS Genet 2008; 4: e1000249.
Imlach WL, Finch SC, Miller JH, Meredith AL, Dalziel JE . A role for BK channels in heart rate regulation in rodents. PLoS One 2010; 5: e8698.
Khaitan D, Sankpal UT, Weksler B, Meister EA, Romero IA, Couraud PO et al. Role of KCNMA1 gene in breast cancer invasion and metastasis to brain. BMC Cancer 2009; 9: 258.
Long X, Tharp DL, Georger MA, Slivano OJ, Lee MY, Wamhoff BR et al. The smooth muscle cell-restricted KCNMB1 ion channel subunit is a direct transcriptional target of serum response factor and myocardin. J Biol Chem 2009; 284: 33671–33682.
Zhang Y, Strissel P, Strick R, Chen J, Nucifora G, Le Beau MM et al. Genomic DNA breakpoints in AML1/RUNX1 and ETO cluster with topoisomerase II DNA cleavage and DNase I hypersensitive sites in t(8;21) leukemia. Proc Natl Acad Sci USA 2002; 99: 3070–3075.
Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M . t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA 1991; 88: 10431–10434.
Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA 2009; 106: 19096–19101.
Hoischen A, Gilissen C, Arts P, Wieskamp N, van DVW et al. Massively parallel sequencing of ataxia genes after array-based enrichment. Hum Mutat 2010; 31: 494–499.
Cohen Jr MM . Perspectives on RUNX genes: an update. Am J Med Genet A 2009; 149A: 2629–2646.
Harada Y, Harada H . Molecular pathways mediating MDS/AML with focus on AML1/RUNX1 point mutations. J Cell Physiol 2009; 220: 16–20.
Agerstam H, Lilljebjorn H, Lassen C, Swedin A, Richter J, Vandenberghe P et al. Fusion gene-mediated truncation of RUNX1 as a potential mechanism underlying disease progression in the 8p11 myeloproliferative syndrome. Genes Chromosomes Cancer 2007; 46: 635–643.
Sun W, Downing JR . Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood 2004; 104: 3565–3572.
Osato M . Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene 2004; 23: 4284–4296.
Gilliland DG . Molecular genetics of human leukemias: new insights into therapy. Semin Hematol 2002; 39: 6–11.
Acknowledgements
We thank H Fiegler and W Haagmans for supporting the initial phase of the study and the array design. We further thank B Kazak for excellent technical assistance and G Schramm, L Du and C Bartenhagen for help on data analysis. This work was supported in part by a grant from Roche Diagnostics GmbH (Penzberg, Germany).
Author contributions
VG and AK designed the study, carried out the experiments, interpreted the data and wrote the manuscript. H-UK and MD performed data analysis. SoS provided technical assistance. FD, SuS, WK, CH and TH provided assistance in the design of the study, characterized patient samples and critically reviewed the manuscript. All authors approved the final version submitted for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
CH, SuS, WK and TH are part owners of the MLL Munich Leukemia Laboratory GmbH. AK, VG, FD and SoS are employed by MLL Munich Leukemia Laboratory GmbH. Other authors declare no conflict of interest. A patent application has been filed under EP09-013670.6.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Grossmann, V., Kohlmann, A., Klein, HU. et al. Targeted next-generation sequencing detects point mutations, insertions, deletions and balanced chromosomal rearrangements as well as identifies novel leukemia-specific fusion genes in a single procedure. Leukemia 25, 671–680 (2011). https://doi.org/10.1038/leu.2010.309
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2010.309
Keywords
This article is cited by
-
Grating-coupled surface-plasmon fluorescence DNA sensor
Applied Physics B (2021)
-
Fast and accurate mutation detection in whole genome sequences of multiple isogenic samples with IsoMut
BMC Bioinformatics (2017)
-
SV-STAT accurately detects structural variation via alignment to reference-based assemblies
Source Code for Biology and Medicine (2016)
-
Positional cloning and next-generation sequencing identified a TGM6 mutation in a large Chinese pedigree with acute myeloid leukaemia
European Journal of Human Genetics (2015)
-
Identification of Multiple Complex Rearrangements Associated with Deletions in the 6q23-27 Region in Sézary Syndrome
Journal of Investigative Dermatology (2013)