Abstract
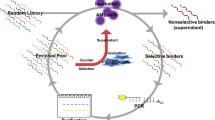
Cell surface proteins can play important roles in cancer pathogenesis. Comprehensive understanding of the surface protein expression patterns of tumor cells and, consequently, the pathogenesis of tumor cells depends on molecular probes against these proteins. To be used effectively for tumor diagnosis, classification and therapy, such probes would be capable of specific binding to targeted tumor cells. Molecular aptamers, designer DNA–RNA probes, can address this challenge by recognizing proteins, peptides and other small molecules with high affinity and specificity. Through a process known as cell-based SELEX, we used live acute myeloid leukemia (AML) cells to select a group of DNA aptamers, which can recognize AML cells with dissociation constants (Kd's) in the nanomolar range. Interestingly, one aptamer (KH1C12) compared with two control cell lines (K562 and NB4) showed significant selectivity to the target AML cell line (HL60) and could recognize the target cells within a complex mixture of normal bone marrow aspirates. The other two aptamers KK1B10 and KK1D04 recognize targets associated with monocytic differentiation. Our studies show that the selected aptamers can be used as a molecular tool for further understanding surface protein expression patterns on tumor cells and thus providing a foundation for effective molecular analysis of leukemia and its subcategories.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Ikeda A, Shankar DB, Watanabe M, Tamanoi F, Moore TB, Sakamoto KM . Molecular targets and the treatment of myeloid leukemia. Mol Genet Metab 2006; 88: 216–224.
Aribi A, Ravandi F, Giles F . Novel agents in acute myeloid leukemia. Cancer J 2006; 12: 77–91.
Shangguan D, Li Y, Tang Z, Cao Z, Cao ZC, Chen HW et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA 2006; 103: 11838–11843.
Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem 2007; 79: 4900–4907.
Labbaye C, Zhang J, Casanova J-L, Lanotte M . Regulation of myeloblastin messenger RNA expression in myeloid leukemia cells treated with all-trans retinoic acid. Blood 1993; 81: 475–481.
Bhatia M, Kirkland JB, Meckling-Gill KA . Monocytic Differentiation of Promyelocytic Leukemia Cells in Response to 1,25-Dihydroxyvitamin D3 Is Independent of Nuclear Receptor Binding. J. Biol. Chem 1995; 270: 15962–15965.
Leupin N, Kuhn A, Hügli B, Grob TJ, Jaggi R, Tobler A et al. Gene expression profiling reveals consistent differences between clinical samples of human leukemia and their model cell lines. Br J Hematol 2006; 135: 520–523.
Olins AL, Hermann H, Lichter P, Olins DE . Retinoic acid differentiation of HL-60 cells promotes cytoskeletal polarization. Exp Cell Res 2000; 254: 130–142.
Marques-Silva VM, De Souza MHO, Teixeira MCL, Arcuri RA, Rumjanek VM . Myeloid leukemia differentiation by phorbol ester and retinoic acid: a practical approach. J Clin Lab Anal 1990; 4: 342–349.
Mollinedo F, Gajate C, Tugores A, Flores I, Naranjo JR . Differences in expression of transcription factor AP-1 in human promyelocytic HL60 cells during differentiation towards macrophages versus granulocytes. Biochem J 1993; 294: 137–144.
Boyd AW, Metcalf D . Induction of differentiation in HL60 leukemia cells: a cell cycle dependent all-or-none event. Leuk Res 1984; 8: 27–43.
Collins JS . The HL60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood 1987; 70: 1233–1244.
Griffin DJ, Larcom P, Schlossman FS . Use of surface markers to identify a subset of acute myelomonocytic leukemia cells with progenitor cell properties. Blood 1983; 62: 1300–1303.
Cerchia L, Ducongé F, Pestourie C, Boulay J, Aissouni Y, Gombert K et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol 2005; 3: 0697–0704.
Zheng X, Ravatn R, Lin Y, Shih W-C, Rabson A, Strair EH et al. Gene expression of TPA induced differentiation in HL60 cells by DNA microarray analysis. Nucleic Acids Res 2002; 30: 4489–4499.
Yang L, Zhao H, Li SW, Ahrens K, Collins C, Eckenrode S et al. Gene expression profiling during all-trans retinoic acid-induced cell differentiation of acute promyelocytic leukemia cells. J Mol Diagn 2003; 5: 212–221.
Chomienne C, Ballerini P, Balitrand N, Daniel MT, Fenaux P, Castaigne S et al. All-trans retinoic acid in acute promyelocytic leukemias. II. In vitro studies: structure-function relationship. Blood 1990; 76: 1710–1717.
Collins SJ, Robertson KA, Mueller L . Retinioc acid-induced granulocytic differentiation of HL60 myeloid leukemia is mediated directly through the retinoic acid receptor (RAR-α). Mol Cell Biol 1990; 10: 2154–2163.
Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. Journal of Proteome Research 2008; 7: 2133–2139.
Mallikaratchy P, Tang Z, Meng L, Shangguan D, Sefah K, Tan W . Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt's Lymphoma cells. Molecular and Cellular Proteomics 2007; 6: 2230–2238.
Acknowledgements
We thank the DNA sequencing core, ICBR at the University of Florida. This work is supported by NIH GM079359 and CA122648, and NSF grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary information
Rights and permissions
About this article
Cite this article
Sefah, K., Tang, Z., Shangguan, D. et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia 23, 235–244 (2009). https://doi.org/10.1038/leu.2008.335
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2008.335
Keywords
This article is cited by
-
The Application of Aptamer and Research Progress in Liver Disease
Molecular Biotechnology (2024)
-
In vitro selection of DNA aptamers against human osteosarcoma
Investigational New Drugs (2022)
-
Development and characterization of a DNA aptamer for MLL-AF9 expressing acute myeloid leukemia cells using whole cell-SELEX
Scientific Reports (2021)
-
Metal–Organic Framework Nanocarriers for Drug Delivery in Biomedical Applications
Nano-Micro Letters (2020)
-
In vitro selections of mammaglobin A and mammaglobin B aptamers for the recognition of circulating breast tumor cells
Scientific Reports (2017)