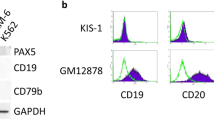
Abstract
A unique feature of the tumor cells (Hodgkin/Reed-Sternberg (HRS)) of classical Hodgkin lymphoma (cHL) is the loss of their B-cell phenotype despite their B-cell origin. Several lines of evidence suggest that epigenomic events, especially promoter DNA methylation, are involved in this silencing of many B-cell-associated genes. Here, we show that DNA demethylation alone or in conjunction with histone acetylation is not able to reconstitute the B-cell-gene expression program in cultured HRS cells. Instead, combined DNA demethylation and histone acetylation of B-cell lines induce an almost complete extinction of their B-cell-expression program and a tremendous upregulation of numerous Hodgkin-characteristic genes, including key players such as Id2 known to be involved in the suppression of the B-cell phenotype. Since the upregulation of Hodgkin-characteristic genes and the extinction of the B-cell-expression program occurred simultaneously, epigenetic changes may also be responsible for the malignant transformation of cHL. The epigenetic upregulation of Hodgkin-characteristic genes thus plays—in addition to promoter DNA hypermethylation of B-cell-associated genes—a pivotal role for the reprogramming of HRS cells and explains why DNA demethylation alone is unable to reconstitute the B-cell-expression program in HRS cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Kanzler H, Kuppers R, Hansmann ML, Rajewsky K . Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med 1996; 184: 1495–1505.
Marafioti T, Hummel M, Foss HD, Laumen H, Korbjuhn P, Anagnostopoulos I et al. Hodgkin and Reed-Sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood 2000; 95: 1443–1450.
Seitz V, Hummel M, Marafioti T, Anagnostopoulos I, Assaf C, Stein H . Detection of clonal T-cell receptor gamma-chain gene rearrangements in Reed-Sternberg cells of classic Hodgkin disease. Blood 2000; 95: 3020–3024.
Falini B, Stein H, Pileri S, Canino S, Farabbi R, Martelli MF et al. Expression of lymphoid-associated antigens on Hodgkin's and Reed-Sternberg cells of Hodgkin's disease. An immunocytochemical study on lymph node cytospins using monoclonal antibodies. Histopathology 1987; 11: 1229–1242.
Schwering I, Brauninger A, Klein U, Jungnickel B, Tinguely M, Diehl V et al. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2003; 101: 1505–1512.
Theil J, Laumen H, Marafioti T, Hummel M, Lenz G, Wirth T et al. Defective octamer-dependent transcription is responsible for silenced immunoglobulin transcription in Reed-Sternberg cells. Blood 2001; 97: 3191–3196.
Doerr JR, Malone CS, Fike FM, Gordon MS, Soghomonian SV, Thomas RK et al. Patterned CpG methylation of silenced B cell gene promoters in classical Hodgkin lymphoma-derived and primary effusion lymphoma cell lines. J Mol Biol 2005; 350: 631–640.
Ushmorov A, Leithauser F, Sakk O, Weinhausel A, Popov SW, Moller P et al. Epigenetic processes play a major role in B-cell-specific gene silencing in classical Hodgkin lymphoma. Blood 2006; 107: 2493–2500.
Mathas S, Janz M, Hummel F, Hummel M, Wollert-Wulf B, Lusatis S et al. Intrinsic inhibition of transcription factor E2A by HLH proteins ABF-1 and Id2 mediates reprogramming of neoplastic B cells in Hodgkin lymphoma. Nat Immunol 2006; 7: 207–215.
Renne C, Martin-Subero JI, Eickernjager M, Hansmann ML, Kuppers R, Siebert R et al. Aberrant expression of ID2, a suppressor of B-cell-specific gene expression, in Hodgkin's lymphoma. Am J Pathol 2006; 169: 655–664.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264.
Kuppers R, Klein U, Schwering I, Distler V, Brauninger A, Cattoretti G et al. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J Clin Invest 2003; 111: 529–537.
Maglott D, Ostell J, Pruitt KD, Tatusova T . Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 2005; 33: D54–D58.
Smyth GK . Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: 1–24, Article 3, e-pub 2004 Feb 12.
Benjamini Y, Hochberg Y . Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 1995; 57: 289–300.
Forsberg EC, Downs KM, Christensen HM, Im H, Nuzzi PA, Bresnick EH . Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc Natl Acad Sci USA 2000; 97: 14494–14499.
Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB . Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 1999; 21: 103–107.
Carbone A, Gloghini A, Aldinucci D, Gattei V, Dalla-Favera R, Gaidano G . Expression pattern of MUM1/IRF4 in the spectrum of pathology of Hodgkin's disease. Br J Haematol 2002; 117: 366–372.
Janz M, Hummel M, Truss M, Wollert-Wulf B, Mathas S, Johrens K et al. Classical Hodgkin lymphoma is characterized by high constitutive expression of activating transcription factor 3 (ATF3), which promotes viability of Hodgkin/Reed-Sternberg cells. Blood 2006; 107: 2536–2539.
Jundt F, Anagnostopoulos I, Forster R, Mathas S, Stein H, Dorken B . Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood 2002; 99: 3398–3403.
Lund R, Ahlfors H, Kainonen E, Lahesmaa AM, Dixon C, Lahesmaa R . Identification of genes involved in the initiation of human Th1 or Th2 cell commitment. Eur J Immunol 2005; 35: 3307–3319.
Hozumi K, Abe N, Chiba S, Hirai H, Habu S . Active form of Notch members can enforce T lymphopoiesis on lymphoid progenitors in the monolayer culture specific for B cell development. J Immunol 2003; 170: 4973–4979.
Kuppers R, Sousa AB, Baur AS, Strickler JG, Rajewsky K, Hansmann ML . Common germinal-center B-cell origin of the malignant cells in two composite lymphomas, involving classical Hodgkin's disease and either follicular lymphoma or B-CLL. Mol Med 2001; 7: 285–292.
Marafioti T, Hummel M, Anagnostopoulos I, Foss HD, Huhn D, Stein H . Classical Hodgkin's disease and follicular lymphoma originating from the same germinal center B cell. J Clin Oncol 1999; 17: 3804–3809.
Acknowledgements
We would like to thank J Oppatt, E Berg and H Lammert for their excellent technical assistance and Professor Dr Franco Gabrielli for providing the anti-DHRS2 (HEP27) antibody. This work was supported by a grant from the Deutsche Forschungsgemeinschaft, Klinische Forschergruppe 105.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary information
Rights and permissions
About this article
Cite this article
Ehlers, A., Oker, E., Bentink, S. et al. Histone acetylation and DNA demethylation of B cells result in a Hodgkin-like phenotype. Leukemia 22, 835–841 (2008). https://doi.org/10.1038/leu.2008.12
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2008.12
Keywords
This article is cited by
-
Sperm DNA methylome abnormalities occur both pre- and post-treatment in men with Hodgkin disease and testicular cancer
Clinical Epigenetics (2023)
-
Insights into the molecular roles of heparan sulfate proteoglycans (HSPGs—syndecans) in autocrine and paracrine growth factor signaling in the pathogenesis of Hodgkin’s lymphoma
Tumor Biology (2016)
-
Primary Mediastinal Large B-Cell Lymphoma, Classic Hodgkin Lymphoma Presenting in the Mediastinum, and Mediastinal Gray Zone Lymphoma: What is the Oncologist To Do?
Current Hematologic Malignancy Reports (2011)
-
Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma
Nature Medicine (2010)
-
Is Hodgkin lymphoma just another B-cell lymphoma?
Current Hematologic Malignancy Reports (2009)