Abstract
In previous studies, a specific paternal lineage with a null value for the Y-chromosome short tandem repeat (Y-STR) marker DYS448 was identified as common among Mongolic- and Turkic-speaking populations. This paternal lineage (temporarily named C3*-DYS448del) was determined to be M217+, M93–, P39–, M48–, M407–, and P53.1–, and its origin and phylogeny remain ambiguous. Here, we analyzed Y-chromosome sequences of 10 male that are related this paternal lineage and redefined it as C3b1a1a1a-F1756 (C3b-F1756). We generated a highly revised phylogenetic tree of haplogroup C3b-F1756, including 21 sub-clades and 360 non-private Y-chromosome polymorphisms. Additionally, we performed a comprehensive analysis of the C3*-DYS448del lineage in eastern Eurasia, including 18 270 samples from 297 populations. Whole Y-chromosome sequences, Y-STR haplotypes, and frequency data were used to generate a distribution map, a network, and age estimations for lineage C3*-DYS448del and its sub-lineages. Considering the historical records of the studied populations, we propose that two major sub-branches of C3b-F1756 may correspond to early expansions of ancestors of modern Mongolic- and Turkic-speaking populations. The large number of newly defined Y-chromosome polymorphisms and the revised phylogenetic tree for C3b-F1756 will assist in investigation of the early history of Altaic-speaking populations in the future.
Similar content being viewed by others
Introduction
Previous studies of the paternal gene pool of Altaic language family populations identified high frequencies of the haplogroup C3*, which were determined to be M217+, M93–, P39–, M48–, M407–, and P53.1–.1, 2, 3, 4, 5 This category contains many different sub-branches of C3*-M217 for which definitive Y-chromosome single nucleotide polymorphism (Y-SNP) markers have yet to be discovered. The majority of C3*-M217 haplogroups belong to the C3*-Star Cluster (currently referred to as C3-F1918), a well-known profile proposed as the paternal lineage of Genghis Khan or his close relatives;3 however, there is another particular lineage among C3*-M217 haplotypes with a null value for the Y-chromosome short tandem repeat (Y-STR) marker DYS448.6, 7 This lineage was temporarily named C3*-DYS448del.
In previous studies, C3*-M217 samples null for DYS448 were mainly obtained from males from Mongolic- and Turkic-speaking populations.4, 5, 6, 7 A small number of studies have investigated the history of the paternal lineage C3*-DYS448del;8 however, its origin and downstream lineages remain ambiguous. In this study, we genotyped Y-SNPs and Y-STRs in additional samples carrying C3*-DYS448del and closely related lineages from eastern Eurasia. Moreover, we sequenced whole Y-chromosomes from 10 samples. The resulting data were used to explore the origin and unique Y-SNP markers of lineage C3*-DYS448del, and to investigate their contributions to the formation of modern Mongolic- and Turkic-speaking populations.
Materials and methods
Blood or saliva samples were collected from unrelated healthy males from populations in eastern Eurasia over the past 10 years. All individuals were adequately informed and signed informed consent forms before their participation. The ethics committee for biological research at the School of Life Sciences in Fudan University approved the study. DNA was extracted from the samples. A number of Y-SNP markers (M130, M217, M93, P39, M48, M407, P53.1, and so on) and 17 STR loci were tested in all DNA samples. Y-chromosome haplogroup frequencies and Y-STR data for haplogroup C-M130 from 297 eastern Eurasian populations were collected from the literature (Supplementary Tables S1 and S2). DNA extracted from 10 selected samples relative to C3*-DYS448del was sent for next-generation sequencing using the Illumina HiSeq2000 platform (San Diego, CA, USA). The details of molecular methods, statistical analysis, workflows for next-generation sequencing, settings for age calculations, nomenclature details, and the final data set for age calculation are provided in Supplementary Text, Supplementary Tables S3 and S4. The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center,9 the Beijing Institute of Genomics (BIG),10 the Chinese Academy of Sciences, under accession number PRJCA000419, and are publicly accessible at http://bigd.big.ac.cn/gsa.
Results
Among 18 270 samples from 297 populations throughout eastern Eurasia, we identified 135 C3*-DYS448del Y-STR haplotypes on 141 samples (Supplementary Tables S1 and S2). The distribution of C3*-DYS448del in eastern Eurasian populations is shown in Figure 1. Generally, the frequencies of this haplotype are low in all studied populations, ranging from 0 to 16.7% (Supplementary Table S1). A distribution peak can be observed in the eastern region of the Mongolian Plateau. Moderate frequencies of C3*-DYS448del were also found in Altai, Teleut, Uzbek, and Kalmyk populations.
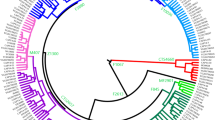
The revised phylogenetic tree for haplogroup C3*-DYS448del contained 21 sub-clades, 360 non-private polymorphisms, and a number of private mutations (Figure 2, see also Supplementary Tables S5 and S6). As shown in Figure 2, haplogroups C3b1a1b-F8535, C3b1a1a3-B77, and C3b1a1a2-P39 are close to C3b1a1a1a-F1756 in the phylogenetic tree. Since all of the sequenced C3*-DYS448del samples had the derived state at marker F1756, we redefined this haplogroup as C3b1a1a1a-F1756 (abbreviated to C3b-F1756). We found that all samples from the Mongolic-speaking populations belonged to sub-branch C3b1a1a1a1a-F3889, while those from the Altai and Hui populations belong to another sub-branch, C3b1a1a1a1b-F8497. Furthermore, two samples from Turkic-speaking populations, FD-Kaz65 and FD-NYG394, also belonged to sub-branch C3b1a1a1a1a-F3889. Additionally, a specific sub-branch C3b1a1a1a1a3-F4022 was found in two samples from Tungisic-speaking populations.
The Y-STR network also provides clues to understanding the internal diversification of lineage C3*-DYS448del. As shown in Figure 3, samples from Altai-kizhi, Kyrgyz, Hui, and Kalmyk populations form a defined clade in the upper right part of the Y-STR network (hereafter referred to as Clade I). Two samples in Clade I from the Hui ethnic group (TJA-033 and TJA-034) were sequenced and form a sub-clade, C3b1a1a1a1b-F8497; however, the relationship among the sequenced samples illustrated as a network (Figure 3) is not entirely consistent with their phylogeny based on Y-SNPs (Figure 2), possibly due to the high mutation rate of Y-STR markers.
The divergence time between haplogroup C3b-F1756 and its most closely related lineage (represented by sample Koryak22071) is ~12 000 years ago (kya).8 The age of the most recent common ancestor (TMRCA) of C3b1a1a1a1-F3830, the major sub-branch of C3b-F1756, was estimated at 4329 years (95% CI, 3538–5 168 years). The ages of the sub-branches of C3b-F1756 are also presented in Supplementary Figure S1. Both the phylogenetic tree and results of age estimation indicate a continuous expansion of C3b-F1756 since ~5.5 kya; however, the frequencies of C3b-F1756 are generally low in modern north Asian populations. This may be caused by recent, very successful, expansions of Mongols.
The cause of the null value of DYS448 in C3b-F1756 samples was investigated. According to sequence data from C3b-F1756 samples, large Y-chromosome deletion of ~0.66 M bp were observed in the region hg19: 24242430–24907270. DYS448 (hg19: 24365070–24365225), DYS589 (hg19: 24485693–24485757), and another nine Y-SNP markers map to this region (Supplementary Tables S5 and S6). Thus, testing of these Y-STR and Y-SNP markers will result in null values in C3b-F1756 samples, and we suggest that special care should be taken when analyzing data in this region from C3*-M217 samples.
Discussion
In this research, we carried out a comprehensive analysis of the paternal lineage C3*-DYS448del in eastern Eurasian populations. We consider that the splitting of two major sub-branches of C3b-F1756, C3b1a1a1a1a-F3889 and C3b1a1a1a1b-F8497, may corresponds to the initial west–east differentiation of the common ancestor group of lineage C3b-F1756. Sub-branch C3b1a1a1a1a-F3889 samples were mainly from modern Mongolic- and Tungusic-speaking populations in the eastern part of the Mongolian Plateau and nearby regions. By contrast, C3b1a1a1a1b-F8497 sub-branch samples were mainly from populations around the Altai Mountain region or regions further west. In addition, the sub-branch C3b1a1a1a1a3-F4022 in Tungisic-speaking populations may represent a particular historical event that is yet to be discovered.
The expansion times for C3b-F1756 and its branch C3b1a1a1a1a-F3889 (~5.5 and 3.3 kya, respectively; Supplementary Figure S1) were much earlier than those for the C3*-Star cluster (~1.1 kya) and C3c-M86 (~2.8 kya).3, 8 We propose that haplogroup C3b-F1756 and its sub-branches may be candidates for the paternal lineages of the ancient Donghu, Xian-Bei, and Shi-Wei tribes who were once the dominant populations in the eastern part of the Mongolian Plateau before the expansion of the Mongols;11 however, more studies of ancient DNA are needed to verify these relationships. The large number of newly defined Y-chromosome polymorphisms and the revised phylogenetic tree of C3b-F1756 generated in this study will be helpful for exploration of the early history of Mongolic- and Turkic-speaking populations in the future.
References
Malyarchuk, B., Derenko, M., Denisova, G., Wozniak, M., Grzybowski, T., Dambueva, I. et al. Phylogeography of the Y-chromosome haplogroup C in northern Eurasia. Ann. Hum. Genet. 74, 539–546 (2010).
Abilev, S., Malyarchuk, B., Derenko, M., Wozniak, M., Grzybowski, T. & Zakharov, I. The Y-chromosome C3* star-cluster attributed to Genghis Khan’s descendants is present at high frequency in the Kerey clan from Kazakhstan. Hum. Biol. 84, 79–89 (2012).
Zerjal, T., Xue, Y., Bertorelle, G., Wells, R. S., Bao, W., Zhu, S. et al. The genetic legacy of the Mongols. Am. J. Hum. Genet. 72, 717–721 (2003).
Malyarchuk, B., Derenko, M., Denisova, G., Khoyt, S., Wozniak, M., Grzybowski, T. et al. Y-chromosome diversity in the Kalmyks at the ethnical and tribal levels. J. Hum. Genet. 58, 804–811 (2013).
Zhong, H., Shi, H., Qi, X. B., Xiao, C. J., Jin, L., Ma, R. Z. et al. Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia. J. Hum. Genet. 55, 428–435 (2010).
Di Cristofaro, J., Pennarun, E., Mazieres, S., Myres, N. M., Lin, A. A., Temori, S. A. et al. Afghan Hindu Kush: where Eurasian sub-continent gene flows converge. PLoS ONE 8, e76748 (2013).
Dulik, M. C., Zhadanov, S. I., Osipova, L. P., Askapuli, A., Gau, L., Gokcumen, O. et al. Mitochondrial DNA and Y chromosome variation provides evidence for a recent common ancestry between Native Americans and Indigenous Altaians. Am. J. Hum. Genet. 90, 229–246 (2012).
Karmin, M., Saag, L., Vicente, M., Wilson Sayres, M. A., Jarve, M., Talas, U. G. et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 25, 459–466 (2015).
Wang, Y., Song, F., Zhu, J., Zhang, S., Yang, Y., Chen, T. et al. GSA: genome sequence archive. Genomics, Proteom. Bioinform. 15, 14–18 (2017).
Members, B. I. G. D. C The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 45, D18–D24 (2017).
Lin, G. A history of Donghu, Inner Mongolian People's Publishing House, Hohhot, (2007).
Acknowledgements
We are grateful to all sample donors. LHW was supported by Future Scientists Project of China Scholarship Council. This work was supported by NSFC for Excellent Young Scholar (nos 31222030, 31671297, 31271338, 31401060), MOE Scientific Research Project (113022A), Ministry of Science and Technology of China (MOST) (2016YFC0900300), and Shanghai Shuguang Project (14SG05).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Wei, LH., Huang, YZ., Yan, S. et al. Phylogeny of Y-chromosome haplogroup C3b-F1756, an important paternal lineage in Altaic-speaking populations. J Hum Genet 62, 915–918 (2017). https://doi.org/10.1038/jhg.2017.60
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.60
This article is cited by
-
Genetic polymorphism of Y-chromosome in Kazakh populations from Southern Kazakhstan
BMC Genomics (2023)
-
Ancient DNA sheds light on the origin and migration patterns of the Xianbei confederation
Archaeological and Anthropological Sciences (2023)
-
Dual origins of the Northwest Chinese Kyrgyz: the admixture of Bronze age Siberian and Medieval Niru’un Mongolian Y chromosomes
Journal of Human Genetics (2022)
-
Origin of ethnic groups, linguistic families, and civilizations in China viewed from the Y chromosome
Molecular Genetics and Genomics (2021)
-
RETRACTED ARTICLE: Y Chromosomal STR haplotypes in Chinese Uyghur, Kazakh and Hui ethnic groups and genetic features of DYS448 null allele and DYS19 duplicated allele
International Journal of Legal Medicine (2021)