Abstract
MicroRNAs (miRNAs) are short non-coding RNAs that regulate the function of target genes at the post-transcriptional phase. miRNAs are considered to have roles in the development, progression and metastasis of cancer. Recent studies have indicated that particular miRNA signatures are correlated with tumor aggressiveness, response to drug therapy and patient outcome in breast cancer. On the other hand, in routine clinical practice, the treatment regimens for breast cancer are determined based on the intrinsic subtype of the primary tumor. Previous studies have shown that miRNA expression profiles of each intrinsic subtypes of breast cancer differ. In hormone receptor-positive/human epidermal growth factor receptor 2 (HER2)-negative breast cancer, miRNA expressions are found to be correlated with endocrine therapy resistance, progesterone receptor expression and heat shock protein activity. Some miRNAs are associated with resistance to HER2-targeted therapy and HER3 expression in HER2-positive breast cancer. In triple-negative breast cancer, miRNA expressions are found to be associated with BRCA mutations, immune system, epithelial–mesenchymal transition, cancer stem cell properties and androgen receptor expression. As it has been clarified that the expression levels and functions of miRNA differ among the various subtypes of breast cancer, and it is necessary to take account of the characteristics of each breast cancer subtype during research into the roles of miRNA in breast cancer. In addition, the discovery of the roles played by miRNAs in breast cancer might provide new opportunities for the development of novel strategies for diagnosing and treating breast cancer.
Similar content being viewed by others
Introduction
Breast cancer exhibits biological heterogeneity in terms of its prognosis and sensitivity to anti-cancer agents. Numerous studies are currently being conducted in an attempt to identify markers of cell growth and differentiation, which are involved in tumor formation and progression, in breast cancer.1
Breast cancer has recently been classified into several intrinsic subtypes, such as the luminal A, luminal B, human epidermal growth factor receptor type 2 (HER2)-enriched, basal-like, claudin-low and normal-like subtypes, based on semi-unsupervised gene expression array analyses.2 In routine clinical practice, these intrinsic subtypes are identified based on a combination of immunohistochemical analyses of estrogen receptor (ER), progesterone receptor (PgR) and HER2 expression, and the Ki67 labeling index (Figure 1), and the following practical classification of intrinsic subtypes was proposed at the St Gallen consensus meeting of breast cancer: luminal A-like type (ER-positive and/or PgR-positive, HER2-negative, low proliferation and low tumor burden), luminal B-like type (ER-positive and/or PgR-positive, HER2-negative, high proliferation and high tumor burden), hormone receptor-positive and HER2-positive type, hormone receptor-negative and HER2-postive type and triple-negative (TN) type (hormone receptor-negative and HER2-negative)3, 4 (Table 1). This classification has provided valuable information about the tumor biology of each subtype and facilitates the appropriate selection of hormonal, chemotherapeutic and HER2-targeting agents during the treatment of breast cancer. The Cancer Genome Atlas Network indicated that the biologic finding of breast cancer subtypes caused by different subsets of genetic and epigenetic abnormalities.1
Immunohistochemical expression levels of the ER, PgR, HER2 and Ki67. (a) Strong ER staining was detected in the cancer cell nuclei. (b) Strong PgR staining was detected in the cancer cell nuclei. (c) The tumor cells exhibited a high Ki67-labeling index. (d) Strong HER2 staining was detected on the cancer cell membrane.
On the other hand, microRNAs (miRNAs) are small (19–22 bases in length) non-coding RNAs, and negatively regulate protein-coding gene expressions by promotion of mRNA degradation or inhibition of translation.5 In breast cancer, various miRNAs have been shown to be deleted or to exhibit downregulated or upregulated expression. Recently, it has been demonstrated that aberrational miRNAs targeting to several cancer-related genes induce cancer initiation, progression, metastasis or drug resistance. In breast cancer, some miRNAs have been shown to upregulate the functions of oncogenes while others stimulate tumor suppressors.6, 7, 8, 9, 10, 11 Previous studies have demonstrated that the miRNA expression profiles of each intrinsic subtype of breast cancer differ.12, 13 In the present review, reports of miRNA research as to the relationship between miRNA expressions and breast cancer subtypes were searched using MEDLINE system and we focus on recent findings regarding the associations between miRNA expressions and the various subtypes of breast cancer.
Biogenesis and functions of miRNA in breast cancer
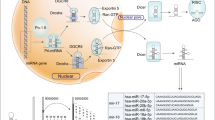
miRNA was initially reported by Lee et al. in 1993,14 and it was identified as a short non-coding RNA with a function of mediating post-transcriptional gene silencing. More than 2500 human miRNAs have subsequently been recorded in the miRBase, a searchable database of published miRNAs.15 miRNAs mediate mRNA degradation and inhibit translation. Most miRNA genes are transcribed by RNA polymerase II in the nucleus, and primary miRNAs (pri-miRNAs) are capped, spliced and polyadenylated.16, 17 Pri-miRNAs are cleaved by a microprocessor composed of the double-stranded RNase III enzyme Drosha and the double-stranded RNA-binding protein DiGeorge syndrome critical region 8 (DGCR8).17, 18, 19 Hairpin-shaped precursor miRNAs (pre-miRNAs)20, 21 are produced from pri-miRNAs by cleavage using Drosha, and they are exported to cytoplasm from nucleus by exportin 5,22, 23, 24 before being further processed by Dicer, an RNase III enzyme that interacts with the 5′ and 3′ ends of pre-miRNA.25, 26 In order to exert their effects, mature miRNAs require ribonucleoprotein complexes, such as RNA-induced silencing complexes, to be assembled.27 The mature single-stranded miRNAs that interact with the Argonaute proteins (AGO1, AGO2, AGO3 and AGO4) in RNA-induced silencing complexes typically bind to the 3′-untranslated regions of their cytosolic mRNA targets. These miRNAs inhibit the translation or promote the deadenylation and degradation of mRNA transcripts.28, 29, 30 This miRNA processing pathway is shown in Figure 2. Overexpressions of oncogenic miRNAs that inhibit tumor suppressor genes are associated with cancer development. On the other hand, reduction or loss of expression of tumor-suppressive miRNAs induce upregulated expression of their target oncogenes.5 In breast cancer, a number of miRNAs have been found to have oncogenic or tumor-suppressive effects, and miRNAs have important roles in tumor initiation, drug resistance and metastasis.6, 7, 8, 9, 10, 11 The function of miRNAs associated with tumor development in breast cancer are shown in Table 2.
Biogenesis and function of microRNA (miRNA). miRNA genes are transcribed by RNA polymerase II (Pol II) in the nucleus to generate primary miRNA (pri-miRNA). Pri-miRNAs are cleaved by a microprocessor composed of a double-stranded RNase III enzyme (Drosha) and the double-stranded RNA-binding protein DiGeorge syndrome critical region 8 (DGCR8). Drosha cleaves pri-miRNA into hairpin-shaped precursor miRNA (pre-miRNA). The pre-miRNAs are then exported from the nucleus to the cytoplasm by exportin 5, before being further processed by Dicer, an RNase III enzyme that interacts with the 5′ and 3′ ends of pre-miRNA. In order to exert their effects, mature miRNA require ribonucleoprotein complexes, such as RNA-induced silencing complexes (RISCs), to be constructed. Mature single-stranded miRNA interact with the Argonaute proteins (AGO) in RISCs to regulate their target genes.
miRNA and intrinsic subtypes of breast cancer
Previous studies have reported that the various breast cancer subtypes exhibit different molecular miRNA signatures.6, 12, 13, 31 Blenkiron et al.13 profiled 309 miRNAs in 93 breast tumors. As a result, they detected different miRNA expression levels between the basal and luminal subtypes. In addition, De Rinaldis et al.32 identified a 46-miRNA signature that could be used to differentiate between breast cancer subtypes. Dvinge et al.33 also obtained similar findings in their research. In a meta-analysis of independent trials, van Schooneveld et al. described various subtype-specific miRNAs. Specifically, let-7c, miR-10a and let-7f were found to be associated with the luminal A type; miR-18a, miR-135b, miR-93 and miR-155 were shown to be associated with the basal type; and miR-142-3p and miR-150 were demonstrated to be associated with the HER2 type.12 Moreover, 453 miRNAs in 29 early-stage breast cancer tumors were profiled, and signatures that could be used to accurately predict the ER, PgR and HER2 status of breast tumor were identified. In addition, miR-342 was expressed most strongly in the ER-positive/HER2-positive tumors.34 MiR-342 influences the ER expression level and the response to tamoxifen.35, 36 MiR-10b, miR-26a and miR-153 have been suggested to be potential biomarkers of triple-negative breast cancer (TNBC).37 miRNAs associated with each subtype of breast cancer are shown in Table 3.
miRNA in hormone receptor-positive/HER2-negative breast cancer
Relationship between miRNA and PgR expression
Approximately 70% of breast cancers are ER-positive and/or PgR-positive. The ER is known to have important roles in the development and progression of breast cancer. It controls the expression of a wide variety of genes and proteins through genomic and non-genomic pathways. In the genomic pathway, estrogen signals are mediated through the ER, which functions as a transcription factor for the signals’ target genes. ER is also activated by signal crosstalk between estrogen and growth factors such as epidermal growth factor and insulin growth factor-1 via transmembrane receptor phosphorylation. PgR expression is induced by the ER, and PgR-related signaling pathways have important roles in the induction, progression and maintenance of neoplastic phenotype in breast cancer. Recent studies have suggested that PgR status needs to be considered when discussing the relative-risk reductions expected from endocrine treatments in individual patients. Prat et al.38 reported that a PgR tumor cell positivity cutoff value of >20% was a significant predictor of the survival differences within luminal-type breast cancers defined by their molecular classification. In addition, we revealed that the extent of PgR expression is a potent prognostic indicator that can aid evaluations of the long-term prognosis of ER-positive/HER2-negative breast cancer.39 PgR and Stat5a are potent prognostic factors of breast cancer and predict the responsiveness of tamoxifen therapy. Finlay-Schultz et al.40 suggested a mechanism by which the progesterone-triggered loss of miR-141 facilitates breast cancer cell de-differentiation through the deregulation of PgR and Stat5a. Furthermore, Lowery et al.34 revealed that four miRNAs (miR-520g, miR-377, miR-527-518a and miR-520f-520c) have the ability to predict PgR status with great accuracy in breast cancer. MiR-29 and miR-513a-5p also influence PgR expression in breast cancer.41
Efficacy of endocrine therapy and miRNA in hormone receptor-positive/HER2-negative breast cancer
miRNAs have important roles in endocrine resistance, and some studies have attempted to identify miRNAs that contribute to the clinical benefits of hormonal therapies. The expression of miR-221 in breast cancer has been identified as a good prognostic marker and is associated with ER positivity and lymph node negativity.42 However, the miR-221/222 cluster is associated with tamoxifen resistance in breast cancer cells.43, 44 Miller et al.45 reported that miR-221/222 expressions were upregulated in endocrine therapy-resistant luminal-type breast cancer cells. MiR-221/222 are negative regulators of p27kip1, a cell cycle inhibitor and tumor suppressor,46, 47, 48, 49, 50 and upregulated expressions of these miRNAs and significant reductions in p27kip1 levels have been reported in tamoxifen-resistant breast cancer cells; therefore, miR-221/222 might regulate tamoxifen sensitivity via the direct targeting of p27kip1.51, 52 Pichiorri et al.53 found that miR-221/222 expressions were modulated by nucleolin at the post-transcriptional level. Recently, it has been indicated that these miRNAs induce resistance to the selective ER downregulator, and this was caused by the activation of β-catenin and the repression of transforming growth factor-β-mediated growth inhibition.54 Lu et al.55 demonstrated that miR-221, miR-222 and miR-181b directly target tissue inhibitor of metalloproteinases (TIMP)3, and MCF7 cells that had been subjected to TIMP3 knockdown were found to be able to grow in the presence of tamoxifen. Furthermore, miR-30c has been identified as an independent predictor of the response to tamoxifen treatment and has been shown to have a role in increasing progression-free survival.56 MiR-301 expression was recently found to be stronger in tumors than in normal tissue, and patients who suffered recurrence after tamoxifen treatment exhibited higher miR-301 levels than those who did not suffer recurrence.57 MiR-375 influences the response to tamoxifen treatment by directly targeting metadherin. The loss of metadherin has been shown to restore sensitivity to endocrine therapy and is correlated with disease-free survival in ER-positive patients.58 He et al.36 demonstrated that miR-342 expression was positively correlated with ER expression and also found that the introduction of miR-342 into estrogen-dependent breast cancer cell lines enhanced their sensitivity to tamoxifen-induced apoptosis. In agreement with this, Cittelly et al.59 reported that the downregulation of miR-342 expression was associated with tamoxifen resistance. miRNAs are also associated with resistance to aromatase inhibitors.60 Masri et al.61 suggested that miR-128a modulates the transforming growth factor-β signaling and survival of letrozole-resistant cell lines. miRNAs expression profiling before and after letrozole treatment reveal post-treatment increases in let-7f expression in both the preclinical and clinical settings.62
miRNA and heat shock proteins in hormone receptor-positive/HER2-negative breast cancer
Heat shock protein (HSP) reveals various effects on multiple oncogenic signaling pathways. The function of HSP is generally considered to be ATP-dependent protein chaperoning. In addition, HSP also has critical roles in post-translational level, which maintains proteins in their correct configurations to ensure their stability and protect carcinoma cells from their apoptosis.63 Overexpressed HSP70 increases levels of unfolded and denatured proteins in stressful cellular conditions. Therefore, HSP70 is considered to be important to maintain the functions of several housekeeping genes. Yiu et al.64 suggested that the downregulation of HSP70 expression was correlated with the treatment response to neoadjuvant endocrine therapy in ER-positive postmenopausal breast cancer patients. The carboxyl terminus of the Hsc70-interacting protein (CHIP) was originally identified as a co-chaperone of E3 ligase, which ubiquitinates misfolded or abnormal proteins presented by molecular chaperones such as HSP70.65 This protein is considered to be a U-box-type ubiquitin ligase that induces the ubiquitination and degradation of its substrates, which include several oncogenic proteins.66, 67 We previously demonstrated that strong CHIP expression is correlated with ER positivity, PgR positivity and HER2 negativity, and identified CHIP expression as a potent prognostic factor in postmenopausal patients with invasive breast cancer.68 Ozgur et al.69 reported that two miRNAs, miR-29a and miR-193b, are associated with breast cancer through their contact with HSP70.
On the other hand, HSP90 is correlated with breast carcinogenesis. HSP90 induces protein folding and refolding under stressful cellular conditions.70 In aromatase inhibitor-resistant breast cancer, growth factor signaling pathways such as insulin-like growth factor-I receptor and HER2-mediated signaling pathways have important functions in tumor growth. Therefore, these pathways have a key role in resistance to aromatase inhibitors and also act as HSP90 client proteins.71 Wong et al.72 suggested that HSP90 inhibitors are effective against aromatase inhibitor-resistant breast cancers. HSP90 is associated with various miRNAs, and miRNA-based inhibition of HSP90 is easier to achieve than miRNA-based HSP70 inhibition.69
miRNA in HER2-positive breast cancer
Efficacy of HER2-targeted therapy and miRNA in HER2-positive breast cancer
Trastuzumab, a humanized monoclonal antibody to HER2 protein, binds to the extracellular domain of HER2 molecules in the cell membrane of carcinoma cells for suppression of HER2 signaling and inhibition of cell proliferation by arresting the cell cycle during the G1 phase. In addition, antibody binding to HER2 leads to antibody-dependent cell-mediated cytotoxicity triggering the carcinoma cell death by immune cells. The treatment strategies for breast cancer changed markedly after trastuzumab was approved as a treatment for HER2-overexpressing breast cancer by the US Food and Drug Administration in 1998. Moreover, other HER2-targeting therapeutic agents such as lapatinib,73 pertuzumab74 and ado-trastuzumab emtansine (T-DM1)75 have been approved as treatments for HER2-overexpressing breast cancer. Jung et al.76 suggested that the plasma miR-210 level is useful for predicting and/or monitoring the therapeutic response to treatments involving trastuzumab, and the upregulation of miR-21 expression has been reported to be associated with trastuzumab resistance in HER2-positive breast cancer.77 In agreement with the latter study, Nishida et al.78 suggested that miR-125a-5p directly targets HER2. MiR-125a-5p was previously shown to strongly suppress the proliferation of gastric cancer cells, and these growth inhibitory effects were enhanced when miR-125a-5p was used in combination with trastuzumab. Ichikawa et al. also found that miR-26a and miR-30b mediate the effects of trastuzumab.79, 80 Furthermore, Iorio et al.81 demonstrated that miR-205, which targets HER3 and impairs the downstream Akt-mediated survival pathway, not only has an oncosuppressive role in breast cancer, but also increases its responsiveness to lapatinib and gefitinib.
Relationship between miRNA and HER3
HER2 is a member of the ErbB-protein family and contributes to a signaling network that operates in the cellular membrane.82 HER2-containing dimers have been shown to enhance downstream signaling.82, 83 As the ligand of HER2 is not discovered, HER2 activation is thought to be strictly dependent on trans-interactions with other members of the HER family such as HER3.81, 84 HER3 has an impaired kinase domain that lacks catalytic function. However, when it forms a heterodimer with a signaling-competent HER family member, HER3 is transphosphorylated and acts as a signaling platform.85
Several studies have demonstrated that HER3 is frequently co-expressed with HER2 in breast cancer, and HER3 has a role in HER2-mediated breast carcinogenesis.86, 87 Iorio et al.81 suggested that the reintroduction of miR-205 into SKBr3 cell increases the responsiveness of lapatinib (tyrosine kinase inhibitors) avoiding HER3-mediated resistance and restoring potent proapoptotic activity. Wang et al.88 predicted that miR-205 binds to the 3′ untranslated regions of HER3 mRNA, and the upregulation of miR-205 reduced HER3 protein expression. Scott et al.89 suggested that the downregulation of HER2 and HER3 protein expression via the overexpression of miR-125a and miR-125b influences the critical features of the malignant cell phenotype, such as proliferative growth, motility and invasiveness in vitro. Wang et al. and Lyu et al. also demonstrated that the overexpression of HER2 promotes HER3 expression via a mechanism involving miR-125a, miR-125b and miR-205 in vivo.90, 91 Bischoff et al. found that miR-148b, miR-149, miR-326, and miR-520a-3p directly reduced HER3 mRNA and protein levels.85 Yan et al.92 provided insights into the role of the miR-143/145 cluster as a tumor suppressor in breast cancer; that is, they suggested that it inhibits HER3 translation in vivo. Yu et al.93 suggested that miR-148a attenuates angiogenesis by inhibiting HER3.
miRNA in triple-negative breast cancer
Molecular subtypes of triple-negative breast cancer
Lehmann et al. revealed that TNBC can be classified into at least six distinct molecular subtypes with differing biological characteristics based on mRNA profiling. These subtypes include two basal-like types (BL1 and BL2), an immunomodulatory type (IM), a mesenchymal type (M), a mesenchymal stem-like type (MSL) and a luminal androgen receptor type (LAR). The BL1 subtype exhibits higher levels of the components of cell division and DNA damage response pathways, including the BRCA pathway. The BL2 subtype has a unique genetic background that involves growth factor signaling (epidermal growth factor, nerve growth factor, MET, Wnt/β-catenin and insulin-like growth factor 1 receptor pathways). The IM subtype displays higher immune cell and immune signal transduction pathway activity. The M and MSL subtypes are both characterized by higher expression levels of the genes involved in motility, the extracellular matrix, cell differentiation pathways and epithelial-to-mesenchymal transition (EMT). Furthermore, the MSL subtype demonstrates elevated cancer stem cell (CSC)-associated gene expression. The LAR subtype is associated with high androgen receptor (AR) expression levels.94, 95 The miRNA expression profiles of these molecular subtypes of TNBC might differ.
Relationship between miRNA and BRCA mutations
The BL subtype of breast cancer is characterized by TNBC and the BL identified by expressions of CK5/6, CK14 and EGFR. Garcia et al. reported that the highest levels of miR-146a and miR-146b-5p were found in BL in vitro and TNBC patients.96 On the other hand, the BRCA1/2 gene is a well-characterized cancer susceptibility gene that is associated with hereditary breast and ovarian cancer.97, 98 Shen et al.99 suggested that genetic polymorphisms in the miR-146a gene might be associated with young age in familial cases of breast or ovarian cancer. The basal phenotype have a strong relationship to BRCA1 mutations, and 80–90% of BRCA1-abnomality expressing cancers exhibiting this phenotype.100 Yan et al.101 reported that sporadic and BRCA1-positive BL subtype cancers demonstrate grade-independent miRNA expression profiles. Furthermore, Shen et al.102 showed that miR-17 binds to BRCA1 mRNA. Chang et al.103,104 discovered that BRCA1 has a role in the epigenetic control of the oncogenic miRNA miR-155. Crippa et al.105 reported that miR-342 regulates BRCA1 expression by modulating the expression of inhibitor of differentiation 4, which in turn negatively regulates BRCA1 expression in breast cancer. Moskwa et al.106 suggested that miR-182 downregulates BRCA1 expression and found that the manipulation of miR-182 expression in breast cell lines affected their sensitivity to poly-ADP ribose polymerase (PARP) 1 inhibition. Tanic et al.107 recently investigated miRNA classifiers in an attempt to predict the BRCA germline mutation status of routinely available formalin-fixed, paraffin-embedded breast tumor biopsy samples based on 6 types of miRNA (miR-142-3p, miR-505, miR-1248, miR-181a-2, miR-25 and miR-340).
Relationship between miRNA and the tumor-associated immune system
The immunity affects all phases of tumor growth from initiation to progression and dissemination. Tumor-infiltrating lymphocytes (TILs) are mononuclear immune cells that infiltrate tumor tissue.108 Several retrospective studies have suggested the possibility of TILs as prognostic factor as well as predict factor of chemotherapy in a part of breast cancer.109 In the Breast International Group 2–98 trial, retrospective–prospective analyses detected a positive correlation between the number of TILs and survival in TNBC.110 The presence of TILs is also associated with increased pathological complete response (pCR) rates. The first randomized controlled trial indicated that a relationship exists between increased numbers of TILs and pCR rates.111 Podshivalova et al.112 suggested that miRNA have an important role in T-lymphocyte activation and also described a mechanism for regulating the impact of miRNA. Jasinski-Bergner reported that natural killer cell and CD8-positive T-lymphocyte ligands are both regulated by the number of miRNAs.113 Therefore, miRNA might contribute to the immune system in breast cancer. Rodriguez et al.114 suggested that bic/miR-155 has a role in regulating homeostasis in the immune system in cancer patients. Zonari et al. reported that miR-155 increases tumor growth by the activation of tumor-associated macrophages in breast cancer. They also found that miR-155 reveals antitumoral effect by acting as an integral effector of immunosurveillance, thereby inhibiting the early stages of breast cancer development.115
On the other hand, programmed cell-death protein 1 and programmed cell death 1 ligand 1 (PD-1/PD-L1) eliminate T-cell activation in various forms of cancer.116 The prospective trials to evaluate the efficacy of antibodies to PD-1/PD-L1 are undergoing in patients with TNBC. These studies may suggest the potential of immune checkpoint inhibitors targeting PD-1/PD-L1 axis in patients with TNBC.117, 118 Iliopoulos et al.119 demonstrated that miR-21 expression was upregulated by ovalbumin stimulation in T cells and also that the inhibition of PD-1 increased miR-21 expression. Chen et al.120 suggested that a relationship exists between miR-200 and PD-L1 expression in human lung cancer. Furthermore, previous studies have indicated that a relationship exists between these immune checkpoints and a number of miRNA; however, these relationships have not yet been elucidated in detail. Further studies are needed in order to determine the relationships between miRNA and the immune system in breast cancer.
Relationship between miRNA and EMT
miRNA might control tumor cell migration caused by EMT and suppress the metastatic potential of breast cancer. Smad and Twist have recently been shown to favor the metastatic dissemination of cancer cells through their abilities to induce EMT. Twist is thought to increase the invasiveness of cancer cell and upregulate miR-10b expression in vitro.121, 122 Moreover, the repression of miR-10b decreases the presence of Twist in the bone metastasis of breast cancer.123 These findings suggest that Twist induces the formation of bone metastasis through a miR-10b-dependent mechanism in breast cancer. Snail is a zinc finger transcriptional repressor, the pathological expression of which has been linked to cancer cell EMT programs and the induction of tissue invasive activity.124, 125, 126, 127 MiR-34a reduces the invasiveness of breast cancer cells by repressing EMT through the Snail pathway.128 Gregory et al. found that all five members of the miRNA-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) and miR-205 was markedly downregulated in cells that had undergone EMT in response to ectopic protein tyrosine phosphatase expression. The enforced expression of members of the miR-200 family were also sufficient to prevent transforming growth factor-β-induced EMT.129 At the cellular level, one of the key events associated with miR-103/107 is the induction of EMT via the downregulation of miR-200 expression.130 Song et al. found that miR-22 triggered EMT, enhanced invasiveness, and promoted metastasis in mouse xenografts. They also demonstrated that miR-22 induced metastatic potential by silencing miR-200 through the targeting of ten-eleven translocation proteins, which are a family of methylcytosine dioxygenases.131 Moreover, Park et al.132 showed that these miRNA cooperatively regulated the expression of the E-cadherin transcriptional repressors ZEB1 and ZEB2, which have previously been implicated in EMT and tumor metastasis. Jiang et al. reported that miR-29 mediated EMT and promoted metastasis in breast cancer.133, 134
Cancer stem cell-related miRNA
Recent studies have suggested that miRNA contribute to tumor initiation by regulating the properties of CSC, including their self-renewal, de-differentiation and drug resistance.5, 9, 135 Han et al.136 demonstrated that the formation of CSC-like cells that were undergoing EMT was associated with the overexpression of hypoxia-inducible factor 1α and that this process was regulated by miR-21. Let-7 has an important role to regulate the function of CSC, because the reduction of let-7 expression inhibits differentiation and maintains proliferation. Therefore, let-7 is a potential molecular marker of CSC and might be a therapeutic target for anti-cancer therapy.137, 138 Several recent studies have suggested that miR-200 family members and their target mRNAs are associated with the properties of CSC. Shimono et al.139 revealed that the expression levels of miRNA-200c-141, miR-200b-200a-429 and miR-183-96-182 were consistently downregulated in CSC of breast cancer. Lim et al.140 suggested that the downregulation of miR-200 may induce the conversion of mammary epithelial cell to a stem cell-like phenotype in breast cancer. Song et al.131 reported that miR-22 modified a crucial epigenetic change, promoted EMT, and induced breast cancer stemness. Takahashi et al.141 identified miR-27b as a key regulator of the generation of breast cancer cells with CSC properties.
Relationship between miRNA and the LAR type
The AR is a nuclear receptor that has a role in the complex network of signaling pathways that regulate cell proliferation in breast cancer.142, 143 In breast cancer, some studies demonstrated that androgenic effects mediated by AR stimulate tumor growth, while other studies suggested that AR-mediated effects inhibit the growth of breast cancer cells.144, 145 Therefore, the mechanisms responsible for the loss of AR expression during breast carcinogenesis remain unclear. Although the loss of AR expression is associated with high nuclear grade and a negative ER, PR and HER2 expression status in breast cancer, the significance of AR expression in human breast cancer has been examined in recent studies into TNBC.146, 147 A number of studies have proposed that a correlation exists between specific miRNA and AR expression. The expression levels of several androgen-inducible miRNA (miR-21, miR-101, miR-125b, miR-221 and miR-222) have been well-characterized in prostate cancer cells, and these miRNA have been found to be involved in the progression to androgen independence.148, 149, 150, 151, 152 MiR-21 has an androgen-responsive element, and is directly upregulated by androgens in prostate cancer cell lines. In addition, miR-21 was expressed at higher levels in AR-positive than in AR-negative prostate cancer cells.148, 149, 151 The roles of miRNA in the regulation of AR expression in breast cancer have been investigated. Nakano et al.153 demonstrated that miR-363 is an androgen-inducible miRNA in breast cancer. However, at present there is limited evidence that a relationship exists between the AR and miRNA in breast cancer.
Conclusions
The present review highlighted recent findings regarding the role of miRNA in breast cancer. Previous studies have revealed that breast cancer comprises several intrinsic subtypes with different molecular profiles, and several miRNAs have important roles to determine and regulate such subtypes. The profiling of miRNA expressions in breast cancer and clarifying molecular mechanisms of breast cancer-specific miRNAs are important future topics for basic and clinical research of breast cancer. The elucidation of the roles played by miRNA in breast cancer has provided new opportunities for the development of strategies for the diagnosis and treatment of cancer. Further biological research into the ability of novel agents to regulate miRNA expression is warranted, and miRNA are expected to become a therapeutic target of treatments for breast cancer.
References
Koboldt, D. C., Fulton, R. S., McLellan, M. D., Schmidt, H., Kalicki-Veizer, J., McMichael, J. F. et al. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Perou, C. M., Sorlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Coates, A. S., Winer, E. P., Goldhirsch, A., Gelber, R. D., Gnant, M., Piccart-Gebhart, M. et al. Tailoring therapies-improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 26, 1533–1546 (2015).
Goldhirsch, A., Winer, E. P., Coates, A. S., Gelber, R. D., Piccart-Gebhart, M., Thürlimann, B. et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 24, 2206–2223 (2013).
Goto, Y., Kurozumi, A., Enokida, H., Ichikawa, T. & Seki, N. Functional significance of aberrantly expressed microRNAs in prostate cancer. Int. J. Urol. 22, 242–252 (2015).
Serpico, D., Molino, L. & Cosimo, S. D. microRNAs in breast cancer development and treatment. Cancer Treat. Rev. 40, 595–604 (2014).
Kayani, M., Kayani, M. A., Malik, F. A. & Faryal, R. Role of miRNAs in breast cancer. Asian Pac. J. Cancer Prev. 12, 3175–3180 (2011).
Mulrane, L., McGee, S. F., Gallagher, W. M. & O’Connor, D. miRNA dysregulation in breast cancer. Cancer Res. 73, 6554–6562 (2013).
Takahashi, R. U., Miyazaki, H. & Ochiya, T. The roles of microRNAs in breast cancer. Cancers (Basel) 7, 598–616 (2015).
Graveel, C. R., Calderone, H. M., Westerhuis, J. J., Winn, M. E. & Sempere, L. F. Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer (Dove Med. Press) 7, 59–79 (2015).
Le Quesne, J. & Caldas, C. Micro-RNAs and breast cancer. Mol. Oncol. 4, 230–241 (2010).
van Schooneveld, E., Wildiers, H., Vergote, I., Vermeulen, P. B., Dirix, L. Y. & Van Laere, S. J. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 17, 21 (2015).
Blenkiron, C., Goldstein, L. D., Thorne, N. P., Spiteri, I., Chin, S. F., Dunning, M. J. et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 8, R214 (2007).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Griffiths-Jones, S., Saini, H. K., van Dongen, S. & Enright, A. J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158 (2008).
Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 (2004).
Lin, S. & Gregory, R. I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333 (2015).
Gregory, R. I., Yan, K. P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004).
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F. & Hannon, G. J. Processing of primary microRNAs by the microprocessor complex. Nature 432, 231–235 (2004).
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003).
Han, J., Lee, Y., Yeom, K. H., Kim, Y. K., Jin, H. & Kim, V. N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027 (2004).
Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E. & Kutay, U. Nuclear export of microRNA precursors. Science 303, 95–98 (2004).
Bohnsack, M. T., Czaplinski, K. & Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10, 185–191 (2004).
Yi, R., Qin, Y., Macara, I. G. & Cullen, B. R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016 (2003).
Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001).
Iorio, M. V. & Croce, C. M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 4, 143–159 (2012).
Eastlack, S. C. & Alahari, S. K. MicroRNA and breast cancer: understanding pathogenesis, improving management. Non-Coding RNA 1, 17–43 (2015).
Hayes, J., Peruzzi, P. P. & Lawler, S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 20, 460–469 (2014).
Ha, M. & Kim, V. N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 15, 509–524 (2014).
Chendrimada, T. P., Gregory, R. I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K. et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 (2005).
van Schooneveld, E., Wouters, M. C., Van der Auwera, I., Peeters, D. J., Wildiers, H., Van Dam, P. A. et al. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 14, R34 (2012).
de Rinaldis, E., Gazinska, P., Mera, A., Modrusan, Z., Fedorowicz, G. M., Burford, B. et al. Integrated genomic analysis of triple-negative breast cancers reveals novel microRNAs associated with clinical and molecular phenotypes and sheds light on the pathways they control. BMC Genomics 14, 643 (2013).
Dvinge, H., Git, A., Gräf, S., Salmon-Divon, M., Curtis, C., Sottoriva, A. et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 497, 378–382 (2013).
Lowery, A. J., Miller, N., Devaney, A., McNeill, R. E., Davoren, P. A., Lemetre, C. et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 11, R27 (2009).
Crippa, E., Lusa, L., De Cecco, L., Marchesi, E., Calin, G. A., Radice, P. et al. miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PLoS ONE 9, e87039 (2014).
He, Y. J., Wu, J. Z., Ji, M. H., Ma, T., Qiao, E. Q., Ma, R. et al. miR-342 is associated with estrogen receptor-alpha expression and response to tamoxifen in breast cancer. Exp. Ther. Med. 5, 813–818 (2013).
Fkih M’hamed, I., Privat, M., Ponelle, F., Penault-Llorca, F., Kenani, A. & Bignon, Y. J. Identification of miR-10b, miR-26a, miR-146a and miR-153 as potential triple-negative breast cancer biomarkers. Cell Oncol. (Dordr) 38, 433–442 (2015).
Prat, A., Cheang, M. C., Martin, M., Parker, J. S., Carrasco, E., Caballero, R. et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol. 31, 203–209 (2013).
Kurozumi, S., Matsumoto, H., Hayashi, Y., Kurosumi, M., Tozuka, K., Kubo, K. et al. Degree of PgR expression is a more potent prognostic factor than Ki67 labeling index in ER-positive and HER2-negative breast cancer. Eur J Cancer 50: abstract s193 (2014).
Finlay-Schultz, J., Cittelly, D. M., Hendricks, P., Patel, P., Kabos, P., Jacobsen, B. M. et al. Progesterone downregulation of miR-141 contributes to expansion of stem-like breast cancer cells through maintenance of progesterone receptor and Stat5a. Oncogene 34, 3676–3687 (2015).
Cochrane, D. R., Jacobsen, B. M., Connaghan, K. D., Howe, E. N., Bain, D. L. & Richer, J. K. Progestin regulated miRNAs that mediate progesterone receptor action in breast cancer. Mol. Cell. Endocrinol. 355, 15–24 (2012).
Hanna, J. A., Wimberly, H., Kumar, S., Slack, F., Agarwal, S. & Rimm, D. L. Quantitative analysis of microRNAs in tissue microarrays by in situ hybridization. Biotechniques 52, 235–245 (2012).
Gan, R., Yang, Y., Zhao, L., Lu, J. & Meng, Q. H. Downregulation of miR-221/222 enhances sensitivity of breast cancer cells to tamoxifen through upregulation of TIMP3. Cancer Gene Ther. 21, 290–296 (2014).
Zhao, J. J., Lin, J., Yang, H., Kong, W., He, L., Ma, X. et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 283, 31079–31086 (2008).
Miller, T. E., Ghoshal, K., Ramaswamy, B., Roy, S., Datta, J., Shapiro, C. L. et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 283, 29897–29903 (2008).
Polyak, K., Lee, M. H., Erdjument-Bromage, H., Koff, A., Roberts, J. M., Tempst, P. et al. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78, 59–66 (1994).
Toyoshima, H. & Hunter, T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78, 67–74 (1994).
Tanaka, T., Tatsuno, I., Noguchi, Y., Uchida, D., Oeda, T., Narumiya, S. et al. Activation of cyclin-dependent kinase 2 (Cdk2) in growth-stimulated rat astrocytes. Geranylgeranylated Rho small GTPase(s) are essential for the induction of cyclin E gene expression. J. Biol. Chem. 273, 26772–26778 (1998).
Koff, A., Giordano, A., Desai, D., Yamashita, K., Harper, J. W., Elledge, S. et al. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science 257, 1689–1694 (1992).
Kurozumi, S., Inoue, K., Takei, H., Matsumoto, H., Kurosumi, M., Horiguchi, J. et al. ER, PgR, Ki67, p27(Kip1), and histological grade as predictors of pathological complete response in patients with HER2-positive breast cancer receiving neoadjuvant chemotherapy using taxanes followed by fluorouracil, epirubicin, and cyclophosphamide concomitant with trastuzumab. BMC Cancer 15, 622 (2015).
le Sage, C., Nagel, R., Egan, D. A., Schrier, M., Mesman, E., Mangiola, A. et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 26, 3699–3708 (2007).
Wei, Y., Lai, X., Yu, S., Chen, S., Ma, Y., Zhang, Y. et al. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 147, 423–431 (2014).
Pichiorri, F., Palmieri, D., Luca, L. D., Consiglio, J., You, J., Rocci, A. et al. In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. J. Exp. Med. 210, 951–968 (2013).
Rao, X., Di Leva, G., Li, M., Fang, F., Devlin, C., Hartman-Frey, C. et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 30, 1082–1097 (2011).
Lu, Y., Roy, S., Nuovo, G., Ramaswamy, B., Miller, T., Shapiro, C. et al. Anti-microRNA-222 (anti-miR-222) and -181B suppress growth of tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein and modulating mitogenic signal. J. Biol. Chem. 286, 42292–42302 (2011).
Rodriguez-Gonzalez, F. G., Sieuwerts, A. M., Smid, M., Look, M. P., Meijier-van Gelder, M. E., de Weerd, V. et al. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res. Treat. 127, 43–51 (2011).
Shi, W., Gerster, K., Alajez, N. M., Tsang, J., Waldron, L., Pintilie, M. et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 71, 2926–2937 (2011).
Ward, A., Balwierz, A., Zhang, J. D., Küblbeck, M., Pawitan, Y., Hielscher, T. et al. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene 32, 1173–1182 (2013).
Cittelly, D. M., Das, P. M., Spoelstra, N. S., Edgerton, S. M., Richer, J. K., Thor, A. D. et al. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol. Cancer 9, 317 (2010).
Hayes, E. L. & Lewis-Wambi, J. S. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 17, 40 (2015).
Masri, S., Liu, Z., Phung, S., Wang, E., Yuan, Y. C. & Chen, S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res. Treat. 124, 89–99 (2010).
Shibahara, Y., Miki, Y., Onodera, Y., Hata, S., Chan, M. S. M., Yiu, C. C. P. et al. Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting CYP19A1. J. Pathol. 227, 357–366 (2012).
Garrido, C., Brunet, M., Didelot, C., Zermati, Y., Schmitt, E. & Kroemer, G. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle 5, 2592–2601 (2006).
Yiu, C. C., Chanplakorn, N., Chan, M. S., Loo, W. T., Chow, L. W., Toi, M. et al. Down-regulation of heat-shock protein 70 (HSP-70) correlated with responsiveness to neoadjuvant aromatase inhibitor therapy in breast cancer patients. Anticancer Res. 30, 3465–3472 (2010).
Ballinger, C. A., Connell, P., Wu, Y., Hu, Z., Thompson, L. J., Yin, L. Y. et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535–4545 (1999).
Connell, P., Ballinger, C. A., Jiang, J., Wu, Y., Thompson, L. J., Höhfeld, J. et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell. Biol. 3, 93–96 (2001).
Ahmed, S. F., Deb, S., Paul, I., Chatterjee, A., Mandal, T., Chatterjee, U. et al. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J. Biol. Chem. 287, 15996–16006 (2012).
Kurozumi, S., Yamaguchi, Y., Hayashi, S., Hiyoshi, H., Suda, T., Gohno, T. et al. Prognostic value of the ubiquitin ligase carboxyl terminus of the Hsc70-interacting protein in postmenopausal breast cancer. Cancer Med (e-pub ahead of print 23 June 2016; doi:10.1002/cam4.780).
Ozgur, A., Tutar, L. & Tutar, Y. Regulation of heat shock proteins by miRNAs in human breast cancer. Microrna 3, 118–135 (2014).
Whitley, D., Goldberg, S. P. & Jordan, W. D. Heat shock proteins: a review of the molecular chaperones. J. Vasc. Surg. 29, 748–751 (1999).
Zagouri, F., Bournakis, E., Koutsoukos, K. & Papadimitriou, C. A. Heat shock protein 90 (hsp90) expression and breast cancer. Pharmaceuticals (Basel) 5, 1008–1020 (2012).
Wong, C. & Chen, S. Heat shock protein 90 inhibitors: new mode of therapy to overcome endocrine resistance. Cancer Res. 69, 8670–8677 (2009).
Geyer, C. E., Forster, J., Lindquist, D., Chan, S., Romieu, C. G., Pienkowski, T. et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N. Engl. J. Med. 355, 2733–2743 (2006).
Baselga, J., Cortés, J., Kim, S. B., Im, S. A., Hegg, R., Im, Y. H. et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N. Engl. J. Med. 366, 109–119 (2012).
Verma, S., Miles, D., Gianni, L., Krop, I. E., Welslau, M., Baselga, J. et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N. Engl. J. Med. 367, 1783–1791 (2012).
Jung, E. J., Santarpia, L., Kim, J., Esteva, F. J., Morretti, E., Buzdar, A. U. et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer 118, 2603–2614 (2012).
Gong, C., Yao, Y., Wang, Y., Liu, B., Wu, W., Chen, J. et al. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J. Biol. Chem. 286, 19127–19137 (2011).
Nishida, N., Mimori, K., Fabbri, M., Yokobori, T., Sudo, T., Tanaka, F. et al. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin. Cancer Res. 17, 2725–2733 (2011).
Ichikawa, T., Sato, F., Terasawa, K., Tsuchiya, S., Toi, M., Tsujimoto, G. et al. Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells. PLoS ONE 7, e31422 (2012).
Wang, S. E. & Lin, R. J. MicroRNA and HER2-overexpressing cancer. Microrna 2, 137–147 (2013).
Iorio, M. V., Casalini, P., Piovan, C., Leva, G. D., Merlo, A., Triulzi, T. et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 69, 2195–2200 (2009).
Citri, A. & Yarden, Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell. Biol. 7, 505–516 (2006).
Graus-Porta, D., Beerli, R. R., Daly, J. M. & Hynes, N. E. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 16, 1647–1655 (1997).
Chiu, C. G., Masoudi, H., Leung, S., Voduc, D. K., Gilks, B., Huntsman, D. G. et al. HER-3 overexpression is prognostic of reduced breast cancer survival: a study of 4046 patients. Ann. Surg. 251, 1107–1116 (2010).
Bischoff, A., Bayerlova, M., Strotbek, M., Schmid, S., Beissbarth, T. & Olayioye, M. A. A global microRNA screen identifies regulators of the ErbB receptor signaling network. Cell. Commun. Signal. 13, 5 (2015).
Sassen, A., Rochon, J., Wild, P., Hartmann, A., Hofstaedter, F., Schwarz, S. et al. Cytogenetic analysis of HER1/EGFR, HER2, HER3 and HER4 in 278 breast cancer patients. Breast Cancer Res. 10, R2 (2008).
Park, Y. H., Jung, H. A., Choi, M. K., Chang, W., Choi, Y. L., Do, I. G. et al. Role of HER3 expression and PTEN loss in patients with HER2-overexpressing metastatic breast cancer (MBC) who received taxane plus trastuzumab treatment. Br. J. Cancer 110, 384–391 (2014).
Wang, Z., Liao, H., Deng, Z., Yang, P., Du, N., Zhanng, Y. et al. miRNA-205 affects infiltration and metastasis of breast cancer. Biochem. Biophys. Res. Commun. 441, 139–143 (2013).
Scott, G. K., Goga, A., Bhaumik, D., Berger, C. E., Sullivan, C. S. & Benz, C. C. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 282, 1479–1486 (2007).
Lyu, H., Huang, J., Edgerton, S. M., Thor, A. D., He, Z. & Liu, B. Increased erbB3 promotes erbB2/neu-driven mammary tumor proliferation and co-targeting of erbB2/erbB3 receptors exhibits potent inhibitory effects on breast cancer cells. Int. J. Clin. Exp. Pathol. 8, 6143–6156 (2015).
Wang, S., Huang, J., Lyu, H., Lee, C. K., Tan, J., Wang, J. et al. Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis. 4, e556 (2013).
Yan, X., Chen, X., Liang, H., Deng, T., Chen, W., Zhang, S. et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol. Cancer 13, 220 (2014).
Yu, J., Li, Q., Xu, Q., Liu, L. & Jiang, B. MiR-148a inhibits angiogenesis by targeting ERBB3. J. Biomed. Res. 25, 170–177 (2011).
Lehmann, B. D., Bauer, J. A., Chen, X., Sanders, M. E., Chakravarthy, A. B., Shyr, Y. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 (2011).
Lehmann, B. D. & Pietenpol, J. A. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J. Pathol. 232, 142–150 (2014).
Garcia, A. I., Buisson, M., Bertrand, P., Rimokh, R., Rouleau, E., Lopez, B. S. et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol. Med. 3, 279–290 (2011).
Fackenthal, J. D. & Olopade, O. I. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat. Rev. Cancer 7, 937–948 (2007).
Ramus, S. J. & Gayther, S. A. The contribution of BRCA1 and BRCA2 to ovarian cancer. Mol. Oncol. 3, 138–150 (2009).
Shen, J., Ambrosone, C. B., DiCioccio, R. A., Odunsi, K., Lele, S. B. & Zhao, H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis 29, 1963–1966 (2008).
Foulkes, W. D., Stefansson, I. M., Chappuis, P. O., Begin, L. R., Goffin, J. R., Wong, N. et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl Cancer Inst. 95, 1482–1485 (2003).
Yan, M., Shield-Artin, K., Byrne, D., Deb, S., Waddell, N., kConFab Investigators et al. Comparative microRNA profiling of sporadic and BRCA1 associated basal-like breast cancers. BMC Cancer 15, 506 (2015).
Shen, J., Ambrosone, C. B. & Zhao, H. Novel genetic variants in microRNA genes and familial breast cancer. Int. J. Cancer 124, 1178–1182 (2009).
Chang, S., Wang, R. H., Akagi, K., Kim, K. A., Martin, B. K., Cavallone, L. et al. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat. Med. 17, 1275–1282 (2011).
Chang, S. & Sharan, S. K. BRCA1 and microRNAs: emerging networks and potential therapeutic targets. Mol. Cells 34, 425–432 (2012).
Crippa, E., Lusa, L., Cecco, L. D., Marchesi, E., Calin, G. A., Radice, P. et al. miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PLoS ONE 9, e87039 (2014).
Moskwa, P., Buffa, F. M., Pan, Y., Panchakshari, R., Gottipati, P., Muschel, R. J. et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 41, 210–220 (2011).
Tanic, M., Yanowski, K., Gómez-López, G., Rodriguez-Pinilla, M. S., Marquez-Rodas, I., Osorio, A. et al. MicroRNA expression signatures for the prediction of BRCA1/2 mutation-associated hereditary breast cancer in paraffin-embedded formalin-fixed breast tumors. Int. J. Cancer 136, 593–602 (2015).
Underwood, J. C. E. Lymphoreticular infiltration in human tumours: prognostic and biological implications: a review. Br. J. Cancer 30, 538–548 (1974).
Savas, P., Salgado, R., Denket, C., Sotiriou, C., Darcy, P. K., Smyth, M. J. et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat. Rev. Clin. Oncol. 13, 228–241 (2016).
Loi, S., Sirtaine, N., Piette, F., Salgado, R., Viale, G., Eenoo, F. V. et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 31, 860–867 (2013).
Denkert, C., Loibl, S., Noske, A., Roller, M., Muller, B. M., Komor, M. et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 28, 105–113 (2010).
Podshivalova, K. & Salomon, D. R. MicroRNA regulation of T-lymphocyte immunity: modulation of molecular networks responsible for T-cell activation, differentiation, and development. Crit. Rev. Immunol. 33, 435–476 (2013).
Jasinski-Bergner, S., Mandelboim, O. & Seliger, B. The role of microRNAs in the control of innate immune response in cancer. J. Natl Cancer Inst. 106, pii: dju257 (2014).
Rodriguez, A., Vigorito, E., Clare, S., Warren, M. V., Couttet, P., Soond, D. R. et al. Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611 (2007).
Zonari, E., Pucci, F., Saini, M., Mazzieri, R., Politi, L. S., Gentner, B. et al. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood 122, 243–252 (2013).
Dong, H., Strome, S. E., Salomao, D. R., Tamura, H., Hirano, F., Files, D. B. et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 8, 793–800 (2002).
Emens, L. A., Braiteh, F. S., Cassier, P., DeLord, J.-P., Eder, J. P., Shen, X. et al. Inhibition of PD-L1 by MPDL3280A leads to clinical activity in patients with metastatic triple-negative breast cancer. Cancer Res. 75, aPD1-6 (2015).
Nanda, R., Chow, L. Q., Dees, E. C., Berger, R., Gupta, S., Geva, R. et al. A Phase Ib study of pembrolizumab (MK-3475) in patients with advanced triple-negative breast cancer. Cancer Res. 75, aS1–09 (2015).
Iliopoulos, D., Kavousanaki, M., Ioannou, M., Boumpas, D. & Verginis, P. The negative costimulatory molecule PD-1 modulates the balance between immunity and tolerance via miR-21. Eur. J. Immunol. 41, 1754–1763 (2011).
Chen, L., Gibbons, D. L., Goswami, S., Cortez, M. A., Ahn, Y. H., Byers, L. A. et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 5, 5241 (2014).
Bourguignon, L. Y. W., Wong, G., Earle, C., Krueger, K. & Spevak, C. C. Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J. Biol. Chem. 285, 36721–36735 (2010).
Ma, L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 12, 210 (2010).
Croset, M., Goehrig, D., Frackowiak, A., Bonnelye, E., Ansieau, S., Puisieux, A. et al. TWIST1 expression in breast cancer cells facilitates bone metastasis formation. J. Bone Miner. Res. 29, 1886–1899 (2014).
Moody, S. E., Perez, D., Pan, T. C., Sarkisian, C. J., Portocarrero, C. P., Sterner, C. J. et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8, 197–209 (2005).
Thiery, J. P., Acloque, H., Huang, R. Y. & Nieto, M. A. Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009).
Vincent, T., Neve, E. P., Johnson, J. R., Kukalev, A., Rojo, F., Albanell, J. et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat. Cell. Biol. 11, 943–950 (2009).
Wu, Y., Deng, J., Rychahou, P. G., Qiu, S., Evers, B. M. & Zhou, B. P. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell 15, 416–428 (2009).
Kim, N. H., Kim, H. S., Li, X. Y., Lee, I., Choi, H. S., Kong, S. E. et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J. Cell. Biol. 195, 417–433 (2011).
Gregory, P. A., Bert, A. G., Paterson, E. L., Barry, S. C., Tsykin, A., Farshid, G. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell. Biol. 10, 593–601 (2008).
Martello, G., Rosato, A., Ferrari, F., Manfrin, A., Cordenonsi, M., Dupont, S. et al. A MicroRNA targeting dicer for metastasis control. Cell 141, 1195–1207 (2010).
Song, S. J., Poliseno, L., Song, M. S., Ala, U., Webster, K., Ng, C. et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 154, 311–324 (2013).
Park, S. M., Gaur, A. B., Lengyel, E. & Peter, M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 (2008).
Jiang, H., Zhang, G., Wu, J. H. & Jiang, C. P. Diverse roles of miR-29 in cancer (review). Oncol. Rep. 31, 1509–1516 (2014).
Gebeshuber, C. A., Zatloukal, K. & Martinez, J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 10, 400–405 (2009).
Schwarzenbacher, D., Balic, M. & Pichler, M. The role of microRNAs in breast cancer stem cells. Int. J. Mol. Sci. 14, 14712–14723 (2013).
Han, M., Wang, Y., Liu, M., Bi, X., Bao, J., Zeng, N. et al. MiR-21 regulates epithelial-mesenchymal transition phenotype and hypoxia-inducible factor-1alpha expression in third-sphere forming breast cancer stem cell-like cells. Cancer Sci. 103, 1058–1064 (2012).
Yu, F., Yao, H., Zhu, P., Zhang, X., Pan, Q., Gong, C. et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 (2007).
Barh, D., Malhotra, R., Ravi, B. & Sindhurani, P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr. Oncol. 17, 70–80 (2010).
Shimono, Y., Zabala, M., Cho, R. W., Lobo, N., Dalerba, P., Qian, D. et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603 (2009).
Lim, Y. Y., Wright, J. A., Attema, J. L., Gregory, P. A., Bert, A. G., Smith, E. et al. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J. Cell. Sci. 126, 2256–2266 (2013).
Takahashi, R. U., Miyazaki, H., Takeshita, F., Yamamoto, Y., Minoura, K., Ono, M. et al. Loss of microRNA-27b contributes to breast cancer stem cell generation by activating ENPP1. Nat. Commun. 6, 7318 (2015).
Yeh, S., Hu, Y. C., Wang, P. H., Xie, C., Xu, Q., Tsai, M. Y. et al. Abnormal mammary gland development and growth retardation in female mice and MCF7 breast cancer cells lacking androgen receptor. J. Exp. Med. 198, 1899–1908 (2003).
Liao, D. J. & Dickson, R. B. Roles of androgens in the development, growth, and carcinogenesis of the mammary gland. J. Steroid Biochem. Mol. Biol. 80, 175–189 (2002).
Doane, A. S., Danso, M., Lal, P., Donaton, M., Zhang, L., Hudis, C. et al. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 25, 3994–4008 (2006).
Birrell, S. N., Bentel, J. M., Hickey, T. E., Ricciardelli, C., Weger, M. A., Horsfall, D. J. et al. Androgens induce divergent proliferative responses in human breast cancer cell lines. J. Steroid Biochem. Mol. Biol. 52, 459–467 (1995).
Penault-Llorca, F. & Viale, G. Pathological and molecular diagnosis of triple-negative breast cancer: a clinical perspective. Ann. Oncol. 23, vi19–vi22 (2012).
Pistelli, M., Caramanti, M., Biscotti, T., Santinelli, A., Pagliacci, A., Lisa, M. D. et al. Androgen receptor expression in early triple-negative breast cancer: clinical significance and prognostic associations. Cancers (Basel) 6, 1351–1362 (2014).
Ribas, J., Ni, X., Haffner, M., Wentzel, E. A., Salmasi, A. H., Chowdhury, W. H. et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 69, 7165–7169 (2009).
Ribas, J. & Lupold, S. E. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle 9, 923–929 (2010).
Shi, X. B., Xue, L., Yang, J., Ma, A. H., Zhao, J., Xu, M. et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc. Natl Acad. Sci. USA 104, 19983–19988 (2007).
Li, T., Li, D., Sha, J., Sun, P. & Huang, Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem. Biophys. Res. Commun. 383, 280–285 (2009).
Cao, P., Deng, Z., Wan, M., Huang, W., Cramer, S. D., Xu, J. et al. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol. Cancer 9, 108 (2010).
Nakano, K., Miki, Y., Hata, S., Ebata, A., Takagi, K., Mcnamara, K. M. et al. Identification of androgen-responsive microRNAs and androgen-related genes in breast cancer. Anticancer Res. 33, 4811–4819 (2013).
Taganov, K. D., Boldin, M. P., Chang, K. J. & Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA 103, 12481–12486 (2006).
Pan, Y. Z., Morris, M. E. & Yu, A. M. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol. Pharmacol. 75, 1374–1379 (2009).
Kovalchuk, O., Filkowski, J., Meservy, J., Ilnytskyy, Y., Tryndyak, V. P., Chekhun, V. F. et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 7, 2152–2159 (2008).
Ivanovska, I., Ball, A. S., Diaz, R. L., Magnus, J. F., Kibukawa, M., Schelter, J. M. et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 28, 2167–2174 (2008).
Liang, Z., Wu, H., Xia, J., Li, Y., Zhang, Y., Huang, K. et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem. Pharmacol. 79, 817–824 (2010).
Zhu, Y., Yu, F., Jiao, Y., Feng, J., Tang, W., Yao, H. et al. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin. Cancer Res. 17, 7105–7115 (2011).
Ma, M. T., He, M., Wang, Y., Jiao, X. Y., Zhao, L., Bai, X. F. et al. MiR-487a resensitizes mitoxantrone (MX)-resistant breast cancer cells (MCF-7/MX) to MX by targeting breast cancer resistance protein (BCRP/ABCG2). Cancer Lett. 339, 107–115 (2013).
Bockhorn, J., Dalton, R., Nwachukwu, C., Huang, S., Prat, A., Yee, K. et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat. Commun. 4, 1393 (2013).
Körner, C., Keklikoglou, I., Bender, C., Wörner, A., Münstermann, E., Wiemann, S. et al. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J. Biol. Chem. 288, 8750–8761 (2013).
Hurst, D. R., Edmonds, M. D., Scott, G. K., Benz, C. C., Vaidya, K. S. & Welch, D. R. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 69, 1279–1283 (2009).
Iorio, M. V., Ferracin, M., Liu, C. G., Veronese, A., Spizzo, R., Sabbioni, S. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070 (2005).
Tavazoie, S. F., Alarcón, C., Oskarsson, T., Padua, D., Wang, Q., Bos, P. D. et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 (2008).
Guo, X., Wu, Y. & Hartley, R. S. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 6, 575–583 (2009).
Guttilla, I. K. & White, B. A. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 284, 23204–23216 (2009).
Chou, J., Lin, J. H., Brenot, A., Kim, J. W., Provot, S. & Werb, Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat. Cell. Biol. 15, 201–213 (2013).
Ma, L., Teruya-Feldstein, J. & Weinberg, R. A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449, 682–688 (2007).
Ryu, S., McDonnell, K., Choi, H., Gao, D., Hahn, M., Joshi, N. et al. Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer Cell 23, 63–76 (2013).
Mattie, M. D., Benz, C. C., Bowers, J., Sensinger, K., Wong, L., Scott, G. K. et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 5, 24 (2006).
Enerly, E., Steinfeld, I., Kleivi, K., Leivonen, S. K., Aure, M. R., Russnes, H. G. et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS ONE 6, e16915 (2011).
Acknowledgements
This study was supported by a grant from the Ministry of Health, Labour, and Welfare of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Kurozumi, S., Yamaguchi, Y., Kurosumi, M. et al. Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J Hum Genet 62, 15–24 (2017). https://doi.org/10.1038/jhg.2016.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.89
This article is cited by
-
Research on biomarkers using innovative artificial intelligence systems in breast cancer
International Journal of Clinical Oncology (2024)
-
Engineered extracellular vesicles: potentials in cancer combination therapy
Journal of Nanobiotechnology (2022)
-
Identification of miRNA biomarkers for breast cancer by combining ensemble regularized multinomial logistic regression and Cox regression
BMC Bioinformatics (2022)
-
MicroRNAs as therapeutic targets in breast cancer metastasis
Drug Delivery and Translational Research (2022)
-
Response to neoadjuvant chemotherapy in breast cancer: do microRNAs matter?
Discover Oncology (2022)