Abstract
Gerodermia osteodysplastica is a recessive segmental progeroid disorder mainly characterized by wrinkled skin, generalized connective tissue weakness, infantile onset osteoporosis and normal intelligence. Coding mutations in GORAB, localized on chromosome 1q24.2, are the cause of this disease. 1q24 deletions underlie a spectrum of disorders with intellectual disability and ear abnormalities as phenotypic hallmarks. Here we report on an individual from Azerbaijan originating from a non-consanguineous couple showing short stature, cutis laxa, frequent fractures, facial dysmorphism, cup-shaped ears and intellectual disability. Sanger sequencing of GORAB revealed the seemingly homozygous missense mutation p.Ser175Phe. This mutation was detected in a heterozygous state in the clinically unaffected mother, but was absent in the healthy father. We performed copy-number investigations by high-resolution array-CGH and PCR approaches and found an ~6 Mb de novo deletion spanning 1q23.3-q24.2 in the affected boy. This novel combination of genetic defects very well explains the phenotype that goes beyond the usual presentation of gerodermia osteodysplastica. Our data provide new insight into the phenotypic spectrum of 1q23-q25 deletions and shows that the combination with another pathogenic allele can lead to more severe clinical manifestations.
Similar content being viewed by others
Introduction
Diseases associated with cutis laxa (CL) are very heterogeneous in terms of the observed phenotypes, the modes of inheritance and the underlying genetic defects.1 The autosomal recessive CL forms are mainly caused by defects affecting components of the mitochondria and the secretory pathway.2, 3, 4, 5 The mitochondrial forms have been shown to result from mutations either in ALDH18A1 or PYCR1.2, 5, 6, 7 Affected individuals show intra-uterine and postnatal growth restriction, a typical progeroid facial appearance, CL, translucent skin and intellectual disability. Often patients with PYCR1 mutations are diagnosed to have gerodermia osteodysplastica (GO, MIM#231070).8, 9 However, GO is due to mutations in GORAB. This gene encodes for an effector of the small GTPases Rab6 and Arf5 and is mainly localized to the Golgi apparatus.3, 10
GO-affected individuals show a progeroid appearance, a pronounced osteoporosis and fractures already in infancy. In contrast to most other forms of CL GO patients do not show any signs of intellectual disability.3, 11 GORAB, located on chromosome 1q24 resides in a region prone to copy-number variability. De novo deletions of different sizes were observed in this region causing an overlapping phenotypic spectrum consisting of growth delay, microcephaly, abnormally shaped ears and intellectual disability.12, 13, 14
In the present study we report on an affected individual with typical signs of GO and further features such as intellectual disability, which are not part of the GO phenotypic spectrum.
Patients and methods
Peripheral blood samples were obtained from the affected individual and his parents. Written informed consent for molecular genetic testing and for publication of photographs from the index patient was obtained from his legal representatives. Sequencing of all exons and the flanking intron regions of GORAB (NM_012463) was performed as described previously.3 The Array-CGH (array-based comparative genomic hybridization) experiment for the index patient II-1 was carried out using a whole-genome 1M oligonucleotide array (Agilent, Santa Clara, CA, USA). The data were analyzed using CytoGenomics v.2.7.8.0 (Agilent) software. Analysis settings: ADM-2; Threshold: 6.0; Window Size: 0.2 Mb; Filter: 5 probes, log2ratio=0.29. Analysis of the exact breakpoint was carried out using endpoint PCRs followed by sequencing. All primer sequences are available on request.
Clinical report
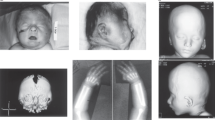
Here we report on a 5-year-old boy from Azerbaijan originating from a non-consanguineous couple (Figure 1a). The index patient was born after an unremarkable pregnancy at 37 weeks of gestation. The mother reported normal birth weight (3 kg, 50th percentile) and length (50 cm, 50–75th percentile). However, the head circumference was described to be abnormal (no data available). Furthermore, a cyanosis was reported that was treated by oxygen supply. He showed generalized CL most pronounced at the hand and feet and bilateral luxation of the hips. In addition, he showed a dysplastic kidney on the right side and undescended testicles. Psychomotor retardation was present. He started to sit with 2 years and to walk with 3 years of life. Speech development started with 7 months using syllables. At the last evaluation with 5 years and 2 months of age he was able to use sentences of three words. He showed short stature (90 cm,<3rd percentile) and the head circumference was 48.5 cm (<3rd percentile). He had a progeroid appearance due to lax and wrinkled skin and reduced subcutaneous fatty tissue. Furthermore, he showed a distinctive facial gestalt with hypertelorism, dysplastic cup-shaped ears, broad nasal bridge and blue sclerae (Figures 1b and c, Table 1). In addition, fifth finger clinodactyly and bilateral broad thumbs were recognizable (Figures 1d and e). He also showed a reduced muscle mass (Figure 1f) and a humerus and a patella fracture were mentioned.
Clinical presentation of the affected individual: (a) Pedigree showing all individuals investigated in the present study. (b, c) Facial appearance of the index patient II-1. Please note the hypertelorism, broad nasal bridge and dysplastic cup-shaped ears. (d, e) He showed a clinodactyly of the fifth finger and broad thumbs bilateral. Furthermore, generalized cutis laxa was most pronounced at the hands and feet. (f) His muscle mass, especially, at the legs was reduced.
Results
Due to the overlapping features with GO, especially in terms of frequent fractures, we performed sequencing of GORAB as described previously.3 We found the nucleotide change c.524C>T causing the amino-acid substitution p.Ser175Phe affecting GORAB in a seemingly homozygous state in the affected individual. Segregation analysis revealed the unaffected mother to be a heterozygous carrier for this alteration. However, in the DNA from the healthy father the variant was absent (Figure 2a). We proved relatedness using STR markers in the DNA from all three individuals (Supplementary Figure 1). Due to this result, we wondered whether a de novo deletion on the second allele of the index patient could be the explanation for the observed homozygosity of the variant c.524C>T. High-resolution array-CGH in the affected individual II-1 revealed a heterozygous deletion on chromosome 1q23-q24 (Figure 2b). By qPCR analysis followed by deletion spanning PCRs (Figure 2c) and subsequent sequencing (Figure 2d), we identified the exact breakpoints between chr1:164037509-170654598 bp (hg19: 1q23.3-q24.2), giving this rearrangement a final size of 6.62 Mb (Figure 2d). Investigation of the parental DNA by qPCR and endpoint PCR revealed that this deletion was only present in the affected individual (Figure 2c, Supplementary Figure 2). These data show that our index patient carries a previously described disease causing GORAB mutation on the maternal allele and a de novo deletion 1q23.3-q24.2 affecting the paternally inherited allele (Figure 2e).
Molecular genetic findings: (a) Sequencing traces of GORAB exon 4 from the index patient and his parents. In the Index patient the alteration c.524C>T is seemingly homozygous, whereas in the unaffected mother this change is heterozygous. In the DNA from the healthy father, the wild-type nucleotide was detected in a homozygous state. (b) Array-CGH profile showing a heterozygous deletion on chromosome 1q23-q24 in the affected individual. (c) Junction fragment analysis of the deleted region on chromosome 1q23.3-q24.2. It was possible to amplify GORAB and PYCR1 exon 3 from all individuals from our index family. The amplification of a junction fragment spanning the breakpoints was only possible in the DNA from the affected individual indicating a de novo occurrence of this particular event. (d) Sequencing trace from the junction fragment showing the exact breakpoint between chr1:164037509-170654598 bp on chromosome 1q23.2-q24.2. (e) Schematic overview showing the combination of the observed mutations in our index patient II-1. A full color version of this figure is available at the Journal of Human Genetics journal online.
Discussion
In the present study we report on a boy with features of GO. However, his clinical appearance was beyond the usual range of this disorder.3, 11 He showed a special facial gestalt with CL, a broad nasal root and dysmorphic, cup-shaped ears. In addition, he showed intellectual disability and microcephaly, what is untypical for GO-affected individuals (Table 1).
The identified mutation p.Ser175Phe affecting GORAB has been described already.10, 15 The GORAB protein affected by this missense mutation is stable, but does not longer localize to the Golgi apparatus. Furthermore, the binding of the mutant protein to the small GTPase Arf5 is altered.10
This underlines the notion that an accurate localization of GORAB at the Golgi is crucial for its biological function. From this point of view, the missense mutation observed also induces a loss of function as all mutations described previously.3 In light of this finding, it is interesting that the observed de novo deletion on chromosome 1q23.3-q24.2 in our index patient contains the GORAB locus. Thus, our index patient shows a biallelic loss of function of GORAB and, additionally, a heterozygous loss of further genetic information from the affected locus.
Chromosome 1q23-q25 has been shown to be prone to de novo deletions of different sizes.13 The clinical spectrum of the 1q23-1q25 de novo deletion syndrome is variable due to the different deletion size, but constant features are proportional growth deficiency, fullness of the upper eyelid, broad nasal bridge, micrognathia, dysplastic ears, small hands and feet, broad thumbs, transverse creases and fifth finger clinodactyly.12, 13 In addition, these patients usually show intellectual disability.13, 14 Our patient shows most of these features due to the detected de novo deletion. However, CL, with reduced subcutaneous fat depots, osteoporosis and recurrent fractures are no features of this deletion syndrome, but well known for gerodermia osteodysplastica. Thus, the observed combination of mutations very well explains the phenotypic presentation of the affected individual.
The combination of a coding mutation in GORAB and a de novo deletion on 1q23.3-q24.2 is, to our knowledge, novel. However, for other diseases like, for example, van den Ende-Gupta syndrome and a 22q11.2 de novo deletion such a combination of two pathogenic events is known.16 Thus, in cases showing a more complex phenotype, which cannot be fully explained by the results of the standard diagnostic procedure, always the existence of a second pathogenic allele in trans should be considered.
In conclusion, in the present study we showed that a de novo deletion on 1q23.3-q24.2 combined with an already known GORAB missense mutation leads to a distinctive phenotype with features from both diseases. Our findings provide new insights into the phenotypic spectrum of 1q23-1q25 deletions and shows that the combination with another pathogenic allele can lead to a more severe clinical presentation.
References
Morava, E., Guillard, M., Lefeber, D. J. & Wevers, R. A. Autosomal recessive cutis laxa syndrome revisited. Eur. J. Hum. Genet. 17, 1099–1110 (2009).
Baumgartner, M. R., Hu, C. A., Almashanu, S., Steel, G., Obie, C., Aral, B. et al. Hyperammonemia with reduced ornithine, citrulline, arginine and proline: a new inborn error caused by a mutation in the gene encoding delta(1)-pyrroline-5-carboxylate synthase. Hum. Mol. Genet. 9, 2853–2858 (2000).
Hennies, H. C., Kornak, U., Zhang, H., Egerer, J., Zhang, X., Seifert, W. et al. Gerodermia osteodysplastica is caused by mutations in SCYL1BP1, a Rab-6 interacting golgin. Nat. Genet. 40, 1410–1412 (2008).
Kornak, U., Reynders, E., Dimopoulou, A., van Reeuwijk, J., Fischer, B., Rajab, A. et al. Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat. Genet. 40, 32–34 (2008).
Reversade, B., Escande-Beillard, N., Dimopoulou, A., Fischer, B., Chng, S. C., Li, Y. et al. Mutations in PYCR1 cause cutis laxa with progeroid features. Nat. Genet. 41, 1016–1021 (2009).
Dimopoulou, A., Fischer, B., Gardeitchik, T., Schroter, P., Kayserili, H., Schlack, C. et al. Genotype-phenotype spectrum of PYCR1-related autosomal recessive cutis laxa. Mol. Genet. Metab. 110, 352–361 (2013).
Fischer-Zirnsak, B., Escande-Beillard, N., Ganesh, J., Tan, Y. X., Al Bughaili, M., Lin, A. E. et al. Recurrent de novo mutations affecting residue Arg138 of pyrroline-5-carboxylate synthase cause a progeroid form of autosomal-dominant cutis laxa. Am. J. Hum. Genet. 97, 483–492 (2015).
Kouwenberg, D., Gardeitchik, T., Wevers, R. A., Haberle, J. & Morava, E. Recognizable phenotype with common occurrence of microcephaly, psychomotor retardation, but no spontaneous bone fractures in autosomal recessive cutis laxa type IIB due to PYCR1 mutations. Am. J. Med. Genet. A 155A, 2331–2332 (2011).
Yildirim, Y., Tolun, A. & Tuysuz, B. The phenotype caused by PYCR1 mutations corresponds to geroderma osteodysplasticum rather than autosomal recessive cutis laxa type 2. Am. J. Med. Genet. A 155A, 134–140 (2011).
Egerer, J., Emmerich, D., Fischer-Zirnsak, B., Chan, W. L., Meierhofer, D., Tuysuz, B. et al. GORAB missense mutations disrupt RAB6 and ARF5 binding and golgi targeting. J. Invest. Dermatol. 135, 2368–2376 (2015).
Rajab, A., Kornak, U., Budde, B. S., Hoffmann, K., Jaeken, J., Nurnberg, P. et al. Geroderma osteodysplasticum hereditaria and wrinkly skin syndrome in 22 patients from Oman. Am. J. Med. Genet. A 146A, 965–976 (2008).
Burkardt, D. D., Rosenfeld, J. A., Helgeson, M. L., Angle, B., Banks, V., Smith, W. E. et al. Distinctive phenotype in 9 patients with deletion of chromosome 1q24-q25. Am. J. Med. Genet. A 155A, 1336–1351 (2011).
Chatron, N., Haddad, V., Andrieux, J., Desir, J., Boute, O., Dieux, A. et al. Refinement of genotype-phenotype correlation in 18 patients carrying a 1q24q25 deletion. Am. J. Med. Genet. A 167A, 1008–1017 (2015).
Mackenroth, L., Hackmann, K., Klink, B., Weber, J. S., Mayer, B., Schrock, E. et al. Interstitial 1q23.3q24.1 deletion in a patient with renal malformation, congenital heart disease, and mild intellectual disability. Am. J. Med. Genet. A 170, 2394–2399 (2016).
Gardeitchik, T., Mohamed, M., Fischer, B., Lammens, M., Lefeber, D., Lace, B. et al. Clinical and biochemical features guiding the diagnostics in neurometabolic cutis laxa. Eur. J. Hum. Genet. 22, 888–895 (2014).
Bedeschi, M. F., Colombo, L., Mari, F., Hofmann, K., Rauch, A., Gentilin, B. et al. Unmasking of a recessive SCARF2 mutation by a 22q11.12 de novo deletion in a patient with Van den Ende-Gupta Syndrome. Mol. Syndromol. 1, 239–245 (2010).
Acknowledgements
We thank the patient and his family for participating in this study. We thank Fabienne Pritsch for excellent technical assistance. This work was funded by the German Federal Ministry of Education and Research (BMBF) (grant 01EC1402B, DIMEOs) and the Elsbeth Bonhoff Stiftung.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Al-Bughaili, M., Neuhann, T., Flöttmann, R. et al. A de novo 1q23.3-q24.2 deletion combined with a GORAB missense mutation causes a distinctive phenotype with cutis laxa. J Hum Genet 62, 325–328 (2017). https://doi.org/10.1038/jhg.2016.111
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.111
This article is cited by
-
mTORC1 controls Golgi architecture and vesicle secretion by phosphorylation of SCYL1
Nature Communications (2022)
-
Narrowing down the region responsible for 1q23.3q24.1 microdeletion by identifying the smallest deletion
Human Genome Variation (2019)