Abstract
Malignant mesothelioma (MM) is an asbestos-related malignancy arising from surface serosal cells of pleural and peritoneal cavities. Somatic mutations of BRCA1 associated protein-1 (BAP1) gene were recently found in MM as well as in uveal melanoma and kidney cancer among the Caucasian and Japanese people. However, frequency of mutations varies among the reported studies, which might be due to presence of undetected gross rearrangements of BAP1 gene that might escape detection by sequencing strategy. We investigated the presence and frequency of gross genomic rearrangements in the BAP1 gene by multiplex ligation-dependent probe amplification (MLPA) in 17 Japanese cases of MM tumors. We found five tumors with partial deletion of BAP1 gene; each tumors displayed partial deletion of exons 1–4 (MM39), exons 1–5 (MM48), exons 11–17 (MM57), exons 1–15 (MM19) and exons 1–16 (MM21). Two tumors (MM34, MM14) had biallelic deletion and four tumors (MM29, MM35, MM45 and MM56) had monoallelic deletion of entire BAP1 gene. Therefore, MLPA analysis revealed large gene rearrangements of BAP1 gene in 65% of MM (11/17). Unusually high frequency of large deletions indicates that the 3p21 chromosomal region surrounding BAP1 gene is structurally unstable. MLPA was useful in characterizing both monoallelic and biallelic deletion of BAP1 gene precisely at exon level.
Similar content being viewed by others
Introduction
Malignant mesothelioma (MM) is highly aggressive neoplasm that arises from the uncontrolled proliferation of surface serosal cells of the pleural, peritoneal and pericardial cavities. MM has been strongly associated with environmental exposure to asbestos.1, 2 However, molecular mechanism of MM remains largely unknown. Somatic mutations in BRCA1 associated protein-1 (BAP1) gene were recently found in 84% of metastasizing uveal melanoma,3 whereas lower frequencies of somatic BAP1 mutation was found in MM in Caucasian people (22, 23, 20 and 36%).4, 5, 6, 7 In Japanese cases, we previously described multiple sequence-level mutations and biallelic deletions of BAP1 gene on seven mesothelioma genome.8 The variable incidence of BAP1 mutations among studies may reflect difference in ethnic background, difference in procedure of tumor cell isolation or difference in methodology of mutation detection.
Most BAP1 mutations described to date are nucleotide-level mutations detected by sequencing strategies, thus, partial or complete gene deletion at exon level might have escaped detection in these strategies. In the present study, we performed multiplex ligation-dependent probe amplification (MLPA) analysis that has been recently used as a highly sensitive method for detection of deletion8, 9 in order to explore frequency and precise nature of genomic rearrangements at exon level affecting BAP1 gene in MM tumors.
Materials and methods
MMs were obtained from the patients diagnosed with MM by pathological examinations at the Hospital of Hyogo College of Medicine as described previously.10 Out of 18 cases studied, sufficient amount of tumor DNA were available in 17 tumors for the present study. ID number of MM tumors classified by histological types are as follows: epitheloid histological type: MM14, MM19, MM21, MM26, MM29, MM34, MM35, MM39, MM45, MM48, MM56, MM57 and MM67; biphasic histological type: MM16, MM30, MM80; and sarcomatoid histological type: MM46. This study was approved by the Ethics Committee of Hyogo College of Medicine and performed in accordance with the Declaration of Helsinki (1995) of the World Medical Association (as revised in Tokyo 2004). All patients provided written informed consent.
MLPA analysis of genomic DNA for each exons of BAP1 gene and data analysis was carried out using MLPA system of MRC-Holland (Amsterdam, The Netherlands) for BAP1 gene. The peak areas achieved using BAP1 specific probes in each sample was first normalized by the average of peak areas achieved by control probes specific for locations from other chromosome. A corresponding calculation was performed for genomic DNA isolated from peripheral blood of 11 individuals and diploid status was defined as the average value from these control DNA. A final ratio dividing the value from the patient samples by the control average value was scored as monoallelic loss when below 0.7 (log2 ratio below −0.52) and biallelic deletion when below 0.25 (log2 ratio below −2).
Results
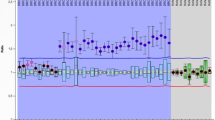
MLPA data for all tumors with partial deletion or biallelic deletion are displayed in Figure 1 (MM39, MM48, MM57, MM19, MM21, MM34 and MM14). Alteration in each tumor is schematically summarized in Figure 2.
MLPA analysis. MLPA data for all 17 exons of BAP1 gene on 3p21.1, as well as those for 10 control probes on chromosome 3p and chromosome 3q are displayed on x axis. Log2 ratio of MLPA data for each probe is indicated on y axis. Panel from top to bottom: MM80, MM39, MM48, MM57, MM19, MM21, MM34 and MM14.
Genomic alteration of 11 MM tumors identified at the BAP1 gene. Region of biallelic deletion, and monoallelic deletion at BAP1 gene identified in 11 MM tumors with rearrangements are shown schematically for each exon of BAP1 gene. Each horizontal thick bar represent genome of the BAP1 gene for MM tumors. ID number of each MM tumors is also indicated along the bar. Top panel shows each exon of BAP1 gene from left to right. A full color version of this figure is available at the Journal of Human Genetics journal online.
Five tumors had partial or interstitial deletion of BAP1 gene; tumor MM39 with partial deletion of exons 1–4, tumor MM48 with partial deletion of exons 1–5, tumor MM57 with partial deletion of exons 11–17, tumor MM19 with partial deletion of exons 1–15 and tumor MM21 with partial deletion of exons 1–16. The remaining allele of BAP1 for each tumor was entirely lost, respectively. Therefore, we were able to precisely characterize gross gene deletions in these tumors at exon level by the use of MLPA that clarified copy number status of each of the 17 exons of BAP1 gene. In addition, tumor MM34 and MM14 had biallelic deletion of entire BAP1 gene. Tumors M29, MM35, MM45 and MM56 had monoallelic deletion of entire BAP1 gene (data not shown). Sequence-level mutations on the remaining allele were described previously among all four tumors with loss of heterozygosity (LOH).10, 11 Together with sequence-level mutations, MLPA revealed BAP1 mutations in 22 out of 34 alleles (65%) of the 17 MM cases that accounts for biallelic inactivation of BAP1 gene in 11 of these tumors (65%). Although MM29 apparently displayed exon 9 deletion by MLPA, it was later found to be caused by failure of MLPA ligation because of 3-bp deletion at MLPA probe site of exon 9 (data not shown).
Discussions
We detected high frequency of somatic gross deletion in the BAP1 gene among 17 Japanese MM tumors and characterized them at exon level. Unusually high frequency of gene rearrangements indicates that the 3p21 chromosomal region surrounding BAP1 gene is structurally unstable. Region of partial deletions observed in BAP1 genes are unique to each tumor and involved region are different among MM tumors, thus no common site of genomic break is apparent on the BAP1 gene.
We previously described association between BAP1 mutation and epitheloid-type among MM in our previous report.10 We here sought any relationship between types of BAP1 mutation and histological types of MM, but no apparent tendency was observed because the number of non-epitheloid-type MM were too small in the present study.
To date, multiple somatic BAP1 mutations have been described in MM tumors, but most of them are limited to nucleotide-level mutations such as non-sense substitutions or small deletions/insertions: 22% (4/18); 23% (12/53); 20% (24/121); and 33% (7/22) among sporadic Caucasian MM tumors by sequencing strategy.4, 5, 6, 7 Yoshikawa et al.10 divided the BAP1 gene into 10 PCR-parts and carried out genomic PCR and nucleotide sequencing of each of the 10 parts with detection of sequence-level mutation and biallelic deletion only on the respective PCR-part of the BAP1 gene. In the present study, we were able to characterize both monoallelic deletion and biallelic deletion of BAP1 gene precisely at every one of the 17 exons that constitute the BAP1 gene by the use of MLPA. Therefore, the significant difference in BAP1 mutation frequencies among the various reports might be due to differences in methodology for detection of mutation. Nasu et al.12 recently described detection of four partial or biallelic deletions of BAP1 gene by MLPA among 22 Caucasian MMs, in addition to some point mutations and splicing mutations. Future investigation in larger panel of MM tumors would be warranted by using comprehensive molecular analysis, that is, sequence analysis, MLPA, immunohistochemisry, to fully understand types and nature of BAP1 gene alteration in MM tumors.13 It is also necessary in the future to carry out the analysis in a much larger set of MM tumors to verify the usefulness of MLPA as a screening method.
References
Carbone, M., Ly, B. H., Dodson, R. F., Pagano, I., Morris, P. T., Dogan, U. A. et al. Malignant mesothelioma: facts, myths, and hypotheses. J. Cell. Physiol. 227, 44–58 (2012).
Baumann, F., Ambrosi, J. P. & Carbone, M. Asbestos is not just asbestos: an unrecognised health hazard. Lancet Oncol. 4, 576–578 (2013).
Harbour, J. W., Onken, M. D., Roberson, E., Duan, S., Cao, L., Worley, L. A. et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413 (2010).
Testa, J. R., Cheung, M., Pei, J., Below, J. E., Tan, Y., Sementino, E. et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat. Genet. 43, 1022–1025 (2011).
Bott, M., Brevet, M., Taylor, B. S., Shimizu, S., Ito, T., Wang, L. et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 43, 668–681 (2011).
Zauderer, M. G., Bott, M., McMillan, R., Sima, C. S., Rusch, V., Krug, L. M. et al. Clinical characteristics of patients with malignant pleural mesothelioma harboring somatic BAP1 mutations. J. Thorac. Oncol. 8, 1430–1433 (2013).
Guo, G., Chmielecki, J., Goparaju, C., Heguy, A., Dolgalev, I., Carbone, M. et al. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 75, 264–269 (2015).
Yamada, H., Shinmura, K., Ito, H., Kasami, M., Sasaki, N., Shima, H. et al. Germline alterations in the CDH1 gene in familial gastric cancer in the Japanese population. Cancer Sci. 102, 1782–1788 (2011).
Cerutti, R., Sahnane, N., Carnevali, I., Furlan, D., Tibiletti, M. G., Chiaravalli, A. M. et al. Identification of the first case of germline duplication of BRCA1 exon 13 in an Italian family. Fam. Cancer 9, 275–282 (2010).
Yoshikawa, Y., Sato, A., Tsujimura, T., Emi, M., Morinaga, T., Fukuoka, K. et al. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Sci. 103, 868–874 (2012).
Yoshikawa, Y., Sato, A., Tsujimura, T., Otsuki, T., Fukuoka, K., Hasegawa, S. et al. Biallelic germline and somatic mutations in malignant mesothelioma: multiple mutations in transcription regulators including mSWI/SNF genes. Int. J. Cancer 136, 560–571 (2015).
Nasu, M., Emi, M., Pastorino, S., Tanji, M., Powers, A., Baumann, F. et al. High incidence of somatic BAP1 alterations in sporadic malignant mesothelioma. J. Thorac. Oncol. 10, 565–576.
Homig-Holzel, C. & Savola, S. Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn. Mol. Pathol. 21, 189–206 (2012).
Acknowledgements
This work was supported in part by a Grant-in-Aid for Scientific Research (KAKENHI, 24590715 to TT, 25460710 to ME) and a Grants-in-Aid for involved researchers.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Emi, M., Yoshikawa, Y., Sato, C. et al. Frequent genomic rearrangements of BRCA1 associated protein-1 (BAP1) gene in Japanese malignant mesothelioma—characterization of deletions at exon level. J Hum Genet 60, 647–649 (2015). https://doi.org/10.1038/jhg.2015.91
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.91