Abstract
Personality is a determinant of behavior and lifestyle associated with health and human diseases. Although personality is known to be a heritable trait, its polygenic nature has made the identification of genetic variants elusive. We performed a genome-wide association study on 1089 Korean women aged 18–40 years whose personality traits were measured with the Revised NEO Personality Inventory for the five-factor model of personality. To reduce environmental factors that may influence personality traits, this study was restricted to young adult women. In the discovery phase, we identified variants of PTPRD (protein tyrosine phosphatase, receptor type D) that associated this gene with the Openness domain. Other genes that were previously reported to be associated with neurological phenotypes were also associated with personality traits. In particular, DRD1 and ORIA2 were linked to Neuroticism, NKAIN2 with Extraversion, HTR5A with Openness and DRD3 with Agreeableness. Data from our replication study of 2090 subjects confirmed the association between ORIA2 and Neuroticism. We first identified and confirmed a novel region on ORIA2 associated with Neuroticism. Candidate genes for psychiatric disorders were also enriched. These findings contribute to our understanding of the genetic architecture of personality traits and provide critical clues to the neurobiological mechanisms that influence them.
Similar content being viewed by others
Introduction
Personality traits are measured with the Revised NEO Personality Inventory (NEO-PI-R), which was designed to characterize the five-factor model (FFM) of the ‘Big Five’ dimensions of personality. The five factors considered are Neuroticism (N), Extraversion (E), Conscientiousness (C), Openness to Experience (O) and Agreeableness (A).1, 2 The five-factor theory asserts that personality traits arise exclusively from biological causes with little or no change in personality traits after early adulthood. Much recent research has examined the effect of gender and age on these five traits. On average, female adults were more neurotic, more extraverted, slightly less open, somewhat more agreeable and slightly more conscientious than males.3 From emerging adulthood through middle age, Neuroticism exhibited a downward trend with age in women, whereas this trait plateaued in men. Extraversion and Openness showed flattened trends, whereas Agreeableness and Conscientiousness exhibited positive trends with age.3, 4, 5, 6
With the development of behavioral genetics, personality was established as a heritable trait. The heritability estimates ranged between 0.40 and 0.60 in family, twin and adoption studies.7, 8, 9 Studies of personality genetics began with the simultaneous publication of two articles in 1996 showing an association between novelty seeking and the D4 dopamine receptor (DRD4) gene.10, 11 This discovery was followed by a report suggesting that activity of the serotonin (5-hydroxytryptamine) transporter protein (SERT or 5-HT) affects the development of anxiety-related traits.12 Two other genes from the serotonin pathway have also been associated with personality, serotonin receptor 2 A (5-HTR2A) and serotonin receptor 2C (5-HT2C).13, 14 However, subsequent studies have been inconsistent in replicating these results, and the literature has been dominated by reports focused on a small number of candidate genes, including DRD and 5-HT.
Recently, the advent of genome-wide association (GWA) studies has had a dramatic impact on the field of personality genetics. Terracciano et al.15 reported the first GWA results of ∼4000 individuals for all five personality dimensions of the FFM, as measured by NEO-PI-R. They found variants associated with the five factors of personality traits, but the effect sizes were small and most of the associations could not be reproduced. Next, the GWA study of personality with Cloninger’s Temperament scales was reported, but genetic variants contributing to personality were not observed.16 A meta-analysis of GWA data for personality traits showed a genome-wide significance, but in silico replication did not confirm any association between the top single nucleotide polymorphisms (SNPs) and personality scores.17, 18 Moreover, recent papers have not reported any evidence to support an association between candidate genes reported previously, including HTR and DRD, and personality.
Considering the effects of gender and age on personality in the design of the population of a genetic study could resolve the uncertainty of the above mentioned results. We present here the results of a GWA study that examined the association between personality traits and genotypes in young Korean women. To minimize the effects of gender and age, a cohort of women within a narrow age group was investigated in this study.
Materials and methods
Participants
Participants were recruited from the Young Women Cohort in Korea, initiated in 2008, which included samples from 2000 Korean women aged 18–40 years whose genotype had been recorded. More than 50 traits were extensively examined through physical examinations and laboratory tests. Within this group, 1140 participants had completed both the personality questionnaire and genotype testing. After completing quality control procedures to eliminate invalid subjects, a total of 1089 participants were included in our GWA analysis. We confirmed though the questionnaire that none of the subjects had received treatment for psychiatric disorders or taken psychoactive drugs.
Personality assessment
Personality traits were assessed using the Korean short version of the original NEO-PI-R,2 a 90-item measure of the five factors of personality (PSI Consulting Corp., Seoul, Korea). The questionnaire consisted of 18 items per factor, Neuroticism (N), Extraversion (E), Openness to experience (O), Agreeableness (A) and Conscientiousness (C). NEO-PI-R has a robust factor structure that has been replicated in Korea19, 20 and in more than >50 cultures.21 Items were answered on a 5-point Likert-type scale ranging from strongly disagree to strongly agree. Phenotype scores for the analysis were computed by summing up the six facets that compose each five factor after reversing negatively keyed items. The NEO-PI-R manual provides a protocol for validity checking based on acquiescence, naysaying and randomness of response.22 On the basis of item-response patterns, 51 subjects were classified as invalid and thus eliminated from this analysis. These subjects provided repetitive answers or had a pattern of acquiescing or naysaying that would have invalidated formal scoring and interpretation of the NEO-PI-R. We did not include missing responses in our data set.
Genotyping and quality control
Genomic DNA was extracted from whole-blood samples using a commercial isolation kit according to the manufacturer’s protocols. The samples were genotyped with the Illumina Human 1 M-Duo DNA Analysis BeadChip (Illumina Inc., San Diego, CA, USA) and BeadStudio software (Illumina Inc.). All sample-wise call rates were 99.43% and average call rate per sample was 99.80%.
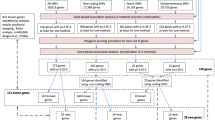
The quality control procedure using PLINK v1.0723 is shown in Figure 1. All analyses were restricted to an SNP on autosomal chromosomes. We evaluated the genotyping completeness of each sample and the SNPs. Any sample missing >5% of its data was excluded. Each sample was also tested to verify that the subject was female. An analysis of identity-by-descent was conducted using PLINK to identify duplicate samples and related individuals. After quality control, 625 112 autosomal SNPs remained, representing 1089 individuals.
SNP imputation
SNP imputation was performed using BEAGLE v3.3.2.24 On the basis of NCBI build 36, we used 90 individuals from Japanese in Tokyo, Japan (JPT), and Han Chinese in Beijing, China (CHB) founders in HapMap as a reference panel comprised of 3.99 million SNPs (HapMap release 23). Only SNPs with an imputation quality score (R2)>0.3 were retained. After removing SNPs with minor allelic frequency <0.05, 2 053 685 imputed SNPs remained for GWA analysis.
GWA analysis: discovery phase
We used the summed values of the six facets of each factor of personality trait as phenotypes in the GWA study. The phenotypes were distributed normally and no transformations were required. GWA analyses were conducted using PLINK. Association analyses were performed on 2 053 685 SNPs and standardized residuals were obtained using a linear regression model for each personality trait and age. We used the additive model in our GWA analyses, and calculated the effective number of independent markers (Me) to adjust for multiple testing, which was performed using the Genetic Type 1 Error Calculator.25 Genetic Type 1 Error Calculator implements the improved Bonferroni correction procedure that estimates Me considering SNPs in linkage disequilibrium (LD). As the estimated Me was 474 906.67 in our data, we assigned the significance and suggestive thresholds of this GWA analysis as P<1.05 × 10−7 (0.05/474 906.67) and P<2.11 × 10−6, respectively.
Replication samples: replication phase
Replication study subjects were selected from an independent cohort from the Kangbuk Samsung Cohort Study, which evaluates natural history, prognosis, and genetic and environmental determinants of a wide range of health traits and diseases among Korean adult men and women. The participants consisted of 2090 young women aged 20–40 years. Personality traits were assessed using the NEO-PI-R, similar to that used in the discovery phase. We selected 40 SNPs among the top 30 ranked SNPs of each factor, which are independent SNPs with low LD (r2<0.5) within the top 30 SNPs. The 40 SNPs were genotyped using the Fluidigm SNPtype Assay (Fluidigm, San Francisco, CA, USA). For four SNPs (rs6660906, rs1411510, rs6475207 and rs16921695), we were unable to design a functioning assay on either platform, therefore alternative SNPs with high LD (r2>0.8) were used: rs6659113, rs964508, rs7852708 and rs1692169, respectively. For quality control, genotyping was performed in duplicate for ∼1% of the samples. Only SNPs that satisfied a concordance rate in duplicate of over 99% and a genotype success rate of over 98% were included in subsequent association analyses.
Statement of ethics
The Institutional Review Board of Ewha Womans University Mokdong Hospital and Kangbuk Samsung Hospital approved this study, and informed consent was obtained from all participants. All applicable institutional and governmental regulations concerning the ethical use of human volunteers were followed during this research.
Results
To search for genetic variants that influence the five factors of personality, we performed a GWA study of 2 053 685 SNPs from 1089 young Korean women. The means of sum and reliability for the five factors are shown in Table 1. Average age of the discovery and replication samples was 26.1 (s.d.=4.6) and 32.9 years (s.d.=3.8), respectively. We included age as a covariate in both analyses. The genomic inflation factor (λ)26 of our GWA analysis was 1.008, 1.000, 1.003, 1.004 and 1.000 for N, E, O, A and C, respectively (Figure 2). The top 30 SNPs with the lowest P-value for each of the five factors are shown in Table 2, which includes only independent SNPs that exhibited a low LD (r2<0.5) within the top 30 SNPs and excludes uncharacterized gene regions. We selected 40 high-ranked SNPs and analyzed them in a replication study of independent samples. Table 2 shows strongest results, and of the 40 SNPs includes only those that are worth discussing. The rest of the SNPs are showed in Supplementary Table S2. Supplementary Table S3 shows the comparison of association results for each trait between Europeans and Koreans by citing results of previous GWA and meta-analysis in Europeans.15, 17 Our results are discussed below.
Quantile–quantile (Q–Q) plots of ∼2 million SNPs from GWA study for five factors of personality traits. The observed P-values (y axis) were compared with the expected P-values under a null distribution (x axis). The black dots represent the observed P-values. The red line indicates the expected line under the null distribution. The gray areas represent the 95% confidence intervals. A full color version of this figure is available at the Journal of Human Genetics journal online.
Neuroticism
Our analysis demonstrated that rs10106540 near the 3′ downstream region of the ST3GAL1 (ST3 beta-galactoside alpha-2,3-sialyltransferase 1) may be associated with Neuroticism (P=4.59 × 10−7). A link between sialyltransferase 4A and bipolar disorder has been reported previously.27 We also identified seven SNPs with P⩽3.33 × 10−5 located near DRD1 on chromosome 5q35 (Figure 3). DRD1 has a role in behavior and psychiatric disorders, including schizophrenia,28 autism,29 attention-deficit-hyperactivity disorder,30 nicotine dependence31 and alcohol dependence.32 Among the top 30 SNPs for Neuroticism, three SNPs (rs265981, P=1.17 × 10−5; rs686, P=1.58 × 10−5 and rs4532, P=1.58 × 10−5, Table 2 and Supplementary Table S1) were previously linked to psychiatric disorders.28, 29, 32 Several SNPs close to olfactory receptor 1A2 (OR1A2) on chromosome 17p13 were also identified, with the strongest signal observed with rs12601685 (P=3.21 × 10−5), which is located 2 kb upstream of OR1A2 (Figure 3). These data are consistent with reports that relate enhanced sensitivity in sensory systems that detect danger to increased neuroticism.33 Our replication study confirmed this result, providing further evidence for an association between OR1A2 and Neuroticism (P=0.05). We also uncovered an association with glypican 6 (GPC6) (Supplementary Tables S1 and S2), which has also been linked with Eysenk Personality Questionnaire—Neuroticism.34 Nevertheless, SNPs previously associated with Neuroticism and reported by Terracciano et al.,15 namely rs362584 in SNAP25 and rs1849710 in TMEM16D, were not linked to this personality trait in our study (Supplementary Table S3).
Regional association plots of SNPs in the GWA analyses for FFM personality. Circles indicate SNPs genotyped in this study, whereas squares indicate SNPs whose genotypes were imputed. Violet dots at each locus indicate the strongest signals detected in the genome-wide scan. SNPs that have an r2 within the top SNP between 0.8 and 1 (red), 0.6 and 0.8 (yellow), 0.4 and 0.6 (green), 0.2 and 0.4 (light blue), and <0.2 (dark blue) are shown. The blue lines show the recombination rates given in the HapMap Phase II JPT+CHB. The genes listed below the plots indicate the RefSeq genes in the loci. The physical positions of SNPs and genes are based on NCBI genome build 36. A full color version of this figure is available at the Journal of Human Genetics journal online.
Extraversion
The highest-ranked association for Extraversion was found at rs6791874 (P=6.9 × 10−7) on chromosome 3 near variants associated with bipolar disorder and attention-deficit-hyperactivity disorder. Unfortunately, the results for every SNP either could not be reproduced or exhibited opposite outcomes. SNP rs2785713 (P=7.1 × 10−6), which maps to the intron of NKAIN2 (Na+/K+-transporting ATPase-interacting 2; Figure 3), showed a consistent effect in the replication study but fell below our P-value threshold for significance. Previous studies associated NKAIN2 with neuroticism and neurological phenotypes.34, 35 SNPs rs17635977 in CDH23 and rs4783307 in CDH13 were not associated with Extraversion, unlike previous reports15 (P=0.40 and P=0.35, respectively).
Openness
SNPs rs2146180, rs10976737 and rs7861684 passed the genome-wide threshold for significance (P<1.05 × 10−7). These SNPs correspond to protein tyrosine phosphatase, receptor type D (PTPRD), which was modestly associated with Persistence (P) in Cloninger’s Temperament.18 We also found moderate association signals with 11 other contiguous SNPs adjacent to PTPRD within the top 30 SNPs for Openness, all with a similar trend (Figure 3). The most significant effect was found in rs2146180 (P=1.7 × 10−8); however, this result failed to replicate (P=0.13). Interestingly, rs1561176, which is located near serotonin receptor 5A (HTR5A), was ranked 21st among the top 30 SNPs (Figure 3) for this trait. The neurotransmitter serotonin (5-HT) has been implicated in a wide range of psychiatric conditions.36, 37 Although rs10251794 in CNTNAP2 was reported in a previous GWA study for Openness,15 rs1477268 and rs2032794 in RASA1, which were identified in a previous GWA study, were not found to be associated with this trait in our study (P=0.33, 0.13 and 0.13, respectively).
Agreeableness
Among the top hits, the biologically notable finding was the association between Agreeableness and SNPs located near DRD3. This receptor is localized to limbic areas of the brain, which are involved in cognitive, emotional and endocrine functions. In the discovery phase, four SNPs (rs2087017, rs10934255, rs1486012 and rs1385884) spanning a region of DRD3 exhibited a high LD (r2>0.8) and an association with this trait (P<9.6 × 10−6; Figure 3). The haplotypes, including SNP rs1486012 located 3′ downstream of the DRD3-coding region, have been associated with schizophrenia.38 DRD3 has also been linked to neuroticism and behavioral inhibition; however, this result has not been reproduced.39, 40 The other highly ranked SNPs for Agreeableness included rs11219218 (P=1.6 × 10−5), which is located 5 kb upstream of SCN3B and is responsible for generating and propagating action potentials in neurons and muscle. We also identified rs11219218 as being significantly associated with this trait (P=0.02); however, the GWA study showed an opposite trend. Finally, rs6832769 in CLOCK, which was linked in a previous GWA analysis to Agreeableness,15 was not associated with this trait in our data (P=0.86).
Conscientiousness
Our GWA data for SNP rs375092 within the intron of IGF2BP3 suggested an association with Conscientiousness (P<4.8 × 10−7), but this result was not confirmed in the replication study. Only one SNP (rs3797269) showed a significant P-value in the replication sample, but the trend was in the opposite direction. In addition to those shown in Table 2, several SNPs in BTAF1 and CPEB3 (cytoplasmic polyadenylation element-binding protein 3) located on chromosome 10q22-23 showed strong associations with Conscientiousness (P<1 × 10−5); however, these results also failed to replicate (Supplementary Tables S1 and S2). Genes located on chromosome 10q22 and 10q23 are associated with psychiatric disorders such as schizophrenia or bipolar disorder. Moreover, chromosome 10 possesses several candidate genes associated with late-onset Alzheimer’s disease.41 SNP rs2576037 in KATNAL2, which has been linked to Conscientiousness in a meta-analysis of GWA,17 was not associated with this trait in our study (P=0.395).
Discussion
We report results from our GWA study of the FFM of personality traits in young Korean women. Thus far, four GWA studies for personality traits have failed to identify any loci with genome-wide significance, although the studies examined up to 4000 subjects. In addition, no variants overlapped in the four studies.15, 16, 42, 43 These results can be attributed to the study design and the sample cohort investigated, which included both genders and broad age range. Gender differences in personality traits have been examined in many recent studies. Combined biological and sociocultural explanations have been offered to explain these findings. Biologically, sex-related differences arise from innate temperamental differences between the sexes that evolved by natural selection.44 Hormonal differences and their effects could also explain gender differences on personality.45 Several studies reported a consistent age difference on personality traits.2, 4 Typically, women declined substantially in Neuroticism throughout adulthood, whereas men declined only modestly.4 To minimize any bias caused by the effects of gender and age, our GWA study included only young adult women. Thus, our results differed from previous GWA studies investigating personality.
ORIA2 showed a strong association with Neuroticism in the discovery and replication phases, making this our most notable finding. Olfactory receptors, which possess a seven-transmembrane domain structure similar to many neurotransmitter and hormone receptors, are responsible for recognizing and transducing G protein-mediated odorant signals. Little research has explored the influence of genetic variation in sensory sensitivity on personality traits, although studies have reported personality differences in visual, auditory, olfactory, tactile and gustatory capacities.33, 46, 47, 48 Mettina et al.49 reported a significant positive relationship between olfactory sensitivity and neuroticism. Another study proposed that people scoring high in emotionality display had higher activation of the limbic system.50 As primary olfactory information processing involves parts of the limbic system, the novel association of ORIA2 with Neuroticism is noteworthy.
We also linked genome-wide significant variations near PTPRD to Openness. However, this finding was not replicated. The protein encoded by PTPRD is a member of the protein tyrosine phosphatase (PTP) family. Studies of similar genes in chicken and fly suggest that PTP is involved in promoting neurite growth and regulating neuronal axon guidance.51, 52 Interestingly, reports have linked PTPRD with Persistence (P) in the meta-analysis of GWAS for TCI of Cloninger’s Temperament.18 A comparative genetic study for NEO and TCI might be informative for researchers interested in personality genetics, although the correlation between Openness and Persistence is not strong according to psychology studies.53
GWA studies and meta-analyses for FFM and Cloninger’s Temperament have been reported previously.15 However, serotonin and dopamine have not been linked to personality traits in these GWA studies. Thus, the association of these genes, namely DRD1, DRD3 and HTR5A, with personality traits and psychiatric disorders in our study is encouraging. Dopamine and serotonin are neurotransmitters that are metabolized by monoamine oxidase. The monoaminergic pathway may have an important role in personality and psychiatric disorders. It is also remarkable that the top 30 SNPs for each factor were enriched with genes associated with neuropsychiatric phenotypes such as schizophrenia, bipolar disorder, attention-deficit-hyperactivity disorder and autism (that is, ST3GAL1,54 PSEN2,55 GPC6,34 NKAIN2,34 ERBB4,56 PDE5A,57 GRIN2A,58 FAM110B59 and ADSL60). Therefore, these genetic variants should be considered further because they may affect personality and facilitate correlation between personality and psychiatric disorders.
Beyond the top 30 SNPs, we also discovered strong signals near CPEB2 and the intron of CPEB3 associated with Extraversion and Conscientiousness, respectively, within the top 50 SNPs (Supplementary Table S1). However, these results could not be reproduced in our replication study. CPEB proteins are crucial for synaptic plasticity and memory in model organisms. Human CPEB3 has reportedly been associated with delayed verbal memory recall, especially emotionally arousing information.61 Other studies also demonstrated an association between personality traits and memory.62 For example, Carlson and Levy63 found an association between Extraversion and short-term memory, whereas Cuttler and Graf64 reported a link between Conscientiousness and prospective memory. Memory, personality, behavior and academic performance are common areas of study for psychologists. Our next challenge is to combine their research with genetics studies so as to advance our understanding of the complexities of personality.
Most associated signals were not replicated in the second stage may be because of the following reasons. First, the sample sizes of our initial and replication populations are underpowered, given that we are testing over 2M SNPs. Secondly, questionnaire-based personality measures lack the reproducibility unlike physical examination or laboratory measures. It has been reported in the literature that social role, life events and social environment are important influences on basic personality traits,4, 65, 66, 67 such as marriage, occupation and parenting, and yet we did not adjust them. The environment factors might have contributed to differences of personality measurements across the samples.
This study represents the first high-density GWA study targeted on FFM personality traits conducted on an East Asian population. Overlap between our GWA results and those previously reported in European-descent populations was not obvious (Supplementary Table S3); however, this may be due to differences in ethnicity, study design or sample size. Each top-associated SNP in analyses of the young women explained an average of 1.7% of variance, which is higher than those reported in other studies and may support a sex-specific effect. Alternatively, this result may overestimate the variance explained by the top SNPs because of the smaller sample size.68 The current GWA method requires a large sample size to identify common variants with small effect size. On the basis of previous GWA studies, personality may also be a polygenic trait affected by many genes with a small effect on complex traits. However, even an SNP with a small effect on personality variance could further extend our understanding of the biology of personality. To increase the power of GWA studies for personality, we must increase the sample size and study different age groups by sex. We found many SNPs enriched in specific loci, but could not prove their association because of only a moderate association with personality traits. However, we may be able to find missing variants by combining moderate genetic effects. Moreover, gene-based and pathway analyses may be alternative methods for discovering missing factors involved in the heritability of personality.
We used a GWA method to understand why individual personality differs, and showed that the key genes associated with personality traits are involved in neurotransmitter signaling and the sensory system with links to psychiatric disorders. Sets of these genes with small effects may influence the development of personality and neurological phenotypes. These results further extend our understanding of the correlation between personality and psychiatric disorders, their genetic architectures and neurobiological effects of personality.
References
McCrae, R. R. & Costa, P. T. Personality in Adulthood: a Five-Factor Theory Perspective 2nd edn, (Guilford Press: New York, NY, USA, 2003).
Costa, P. & McCrae, R. Professional Manual: Revised NEO Personality Inventory (NEO-PI-R) and NEO Five-Factor Inventory (NEO-FFI) Professional Manual, (Psychological Assessment Resources: Odessa: Odessa, FL, USA, 1992).
Soto, C. J., John, O. P., Gosling, S. D. & Potter, J. Age Differences in Personality Traits From 10 to 65: Big Five Domains and Facets in a Large Cross-Sectional Sample. J. Pers. Soc. Psychol. 100, 330–348 (2011).
Srivastava, S., John, O. P., Gosling, S. D. & Potter, J. Development of personality in early and middle adulthood: set like plaster or persistent change? J. Pers. Soc. Psychol. 84, 1041–1053 (2003).
Allemand, M., Zimprich, D. & Hendriks, A. A. J. Age differences in five personality domains across the life span. Dev. Psychol. 44, 758–770 (2008).
Roberts, B. W., Walton, K. E. & Viechtbauer, W. Patterns of mean-level change in personality traits across the life course: A meta-analysis of longitudinal studies. Psychol. Bull. 132, 1–25 (2006).
Horn, J. M., Plomin, R. & Rosenman, R. Heritability of personality traits in adult male twins. Behav. Genet. 6, 17–30 (1976).
Floderus-Myrhed, B., Pedersen, N. & Rasmuson, I. Assessment of heritability for personality, based on a short-form of the Eysenck Personality Inventory: a study of 12,898 twin pairs. Behav. Genet. 10, 153–162 (1980).
Jang, K. L., Livesley, W. J. & Vernon, P. A. Heritability of the big five personality dimensions and their facets: a twin study. J. Pers. 64, 577–591 (1996).
Benjamin, J., Li, L., Patterson, C., Greenberg, B. D., Murphy, D. L. & Hamer, D. H. Population and familial association between the D4 dopamine receptor gene and measures of Novelty Seeking. Nat. Genet. 12, 81–84 (1996).
Ebstein, R. P., Novick, O., Umansky, R., Priel, B., Osher, Y., Blaine, D. et al. Dopamine D4 receptor (D4DR) exon III polymorphism associated with the human personality trait of Novelty Seeking. Nat. Genet. 12, 78–80 (1996).
Lesch, K. P., Bengel, D., Heils, A., Sabol, S. Z., Greenberg, B. D., Petri, S. et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274, 1527–1531 (1996).
Alfimova, M. V., Monakhov, M. V., Golimbet, V. E., Korovaitseva, G. I. & Lyashenko, G. L. Analysis of associations between 5-HTT, 5-HTR2A, and GABRA6 gene polymorphisms and health-associated personality traits. Bull. Exp. Biol. Med. 149, 434–436 (2010).
Ebstein, R. P., Segman, R., Benjamin, J., Osher, Y., Nemanov, L. & Belmaker, R. H. 5-HT2C (HTR2C) serotonin receptor gene polymorphism associated with the human personality trait of reward dependence: interaction with dopamine D4 receptor (D4DR) and dopamine D3 receptor (D3DR) polymorphisms. Am. J. Med. Genet. 74, 65–72 (1997).
Terracciano, A., Sanna, S., Uda, M., Deiana, B., Usala, G., Busonero, F. et al. Genome-wide association scan for five major dimensions of personality. Mol. Psychiatry 15, 647–656 (2010).
Verweij, K. J., Zietsch, B. P., Medland, S. E., Gordon, S. D., Benyamin, B., Nyholt, D. R. et al. A genome-wide association study of Cloninger's temperament scales: implications for the evolutionary genetics of personality. Biol. Psychol. 85, 306–317 (2010).
de Moor, M. H., Costa, P. T., Terracciano, A., Krueger, R. F., de Geus, E. J., Toshiko, T. et al. Meta-analysis of genome-wide association studies for personality. Mol. Psychiatry 17, 337–349 (2012).
Service, S. K., Verweij, K. J., Lahti, J., Congdon, E., Ekelund, J., Hintsanen, M. et al. A genome-wide meta-analysis of association studies of Cloninger's Temperament Scales. Transl. Psychiatry 2, e116 (2012).
Ahn, C. K. & Chae, J. H. Standardization of the Korean version of the Revised NEO Personality Inventory. Korean J. Counsel. Psychother. 9, 443–473 (1997).
Piedmont, R. L. & Chae, J. H. Cross-cultural generalizability of the Five-Factor Model of personality: development and validation of the NEO-PI-R for Koreans. J. Cross. Cult. Psychol. 28, 131–155 (1997).
McCrae, R. R. & Terracciano, A. Personality Profiles of Cultures P. Universal features of personality traits from the observer's perspective: data from 50 cultures. J. Pers. Soc. Psychol. 88, 547–561 (2005).
Costa, P. & McCrae, R. Trait Psychology Comes of Age, University of Nebraska Press: Lincoln Nebraska Symp on Motivation 1991 (1992).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Browning, B. L. & Browning, S. R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84, 210–223 (2009).
Li, M. X., Yeung, J. M., Cherny, S. S. & Sham, P. C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 131, 747–756 (2012).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Perlis, R. H., Purcell, S., Fagerness, J., Kirby, A., Petryshen, T. L., Fan, J. et al. Family-based association study of lithium-related and other candidate genes in bipolar disorder. Arch. Gen. Psychiatry 65, 53–61 (2008).
Zhu, F., Yan, C. X., Wang, Q., Zhu, Y. S., Zhao, Y., Huang, J. et al. An association study between dopamine D1 receptor gene polymorphisms and the risk of schizophrenia. Brain. Res. 1420, 106–113 (2011).
Hettinger, J. A., Liu, X., Schwartz, C. E., Michaelis, R. C. & Holden, J. J. A DRD1 haplotype is associated with risk for autism spectrum disorders in male-only affected sib-pair families. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 147B, 628–636 (2008).
Gao, X. P., Su, L. Y., Zhao, A. L., Luo, X. R. & Xia, K. [Association of 14 polymorphisms in the five candidate genes and attention deficit hyperactivity disorder]. Zhongguo dang dai er ke za zhi 11, 617–622 (2009).
Huang, W., Ma, J. Z., Payne, T. J., Beuten, J., Dupont, R. T. & Li, M. D. Significant association of DRD1 with nicotine dependence. Hum. Genet. 123, 133–140 (2008).
Batel, P., Houchi, H., Daoust, M., Ramoz, N., Naassila, M. & Gorwood, P. A haplotype of the DRD1 gene is associated with alcohol dependence. Alcohol. Clin. Exp. Res. 32, 567–572 (2008).
Croy, I., Springborn, M., Lotsch, J., Johnston, A. N. & Hummel, T. Agreeable smellers and sensitive neurotics—correlations among personality traits and sensory thresholds. PLoS One 6, e18701 (2011).
Calboli, F. C., Tozzi, F., Galwey, N. W., Antoniades, A., Mooser, V., Preisig, M. et al. A genome-wide association study of neuroticism in a population-based sample. PLoS One 5, e11504 (2010).
Bocciardi, R., Giorda, R., Marigo, V., Zordan, P., Montanaro, D., Gimelli, S. et al. Molecular characterization of a t(2;6) balanced translocation that is associated with a complex phenotype and leads to truncation of the TCBA1 gene. Hum. Mutat. 26, 426–436 (2005).
Terracciano, A., Tanaka, T., Sutin, A. R., Deiana, B., Balaci, L., Sanna, S. et al. BDNF Val66Met is associated with introversion and interacts with 5-HTTLPR to influence neuroticism. Neuropsychopharmacology 35, 1083–1089 (2010).
Yosifova, A., Mushiroda, T., Stoianov, D., Vazharova, R., Dimova, I., Karachanak, S. et al. Case-control association study of 65 candidate genes revealed a possible association of a SNP of HTR5A to be a factor susceptible to bipolar disease in Bulgarian population. J. Affect. Disord. 117, 87–97 (2009).
Dominguez, E., Loza, M. I., Padin, F., Gesteira, A., Paz, E., Paramo, M. et al. Extensive linkage disequilibrium mapping at HTR2A and DRD3 for schizophrenia susceptibility genes in the Galician population. Schizophr. Res. 90, 123–129 (2007).
Staner, L., Hilger, C., Hentges, F., Monreal, J., Hoffmann, A., Couturier, M. et al. Association between novelty-seeking and the dopamine D3 receptor gene in bipolar patients: a preliminary report. Am. J. Med. Genet. 81, 192–194 (1998).
Henderson, A. S., Korten, A. E., Jorm, A. F., Jacomb, P. A., Christensen, H., Rodgers, B. et al. COMT and DRD3 polymorphisms, environmental exposures, and personality traits related to common mental disorders. Am. J. Med. Genet. 96, 102–107 (2000).
Grupe, A., Li, Y., Rowland, C., Nowotny, P., Hinrichs, A. L., Smemo, S. et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am. J. Hum. Genet. 78, 78–88 (2006).
Shifman, S., Bhomra, A., Smiley, S., Wray, N. R., James, M. R., Martin, N. G. et al. A whole genome association study of neuroticism using DNA pooling. Mol. Psychiatry 13, 302–312 (2008).
van den Oord, E. J., Kuo, P. H., Hartmann, A. M., Webb, B. T., Moller, H. J., Hettema, J. M. et al. Genomewide association analysis followed by a replication study implicates a novel candidate gene for neuroticism. Arch. Gen. Psychiatry 65, 1062–1071 (2008).
Costa, P. T. Jr, Terracciano, A. & McCrae, R. R. Gender differences in personality traits across cultures: robust and surprising findings. J. Pers. Soc. Psychol. 81, 322–331 (2001).
Berenbaum, S. A. Effects of early androgens on sex-typed activities and interests in adolescents with congenital adrenal hyperplasia. Horm. Behav. 35, 102–110 (1999).
Hollis, J., Allen, P. M., Fleischmann, D. & Aulak, R. Personality dimensions of people who suffer from visual stress. Ophthalmic Physiol. Opt. 27, 603–610 (2007).
Welge-Lussen, A. Ageing, neurodegeneration, and olfactory and gustatory loss. B-Ent 5 (Suppl 13), 129–132 (2009).
Fillingim, R. B., King, C. D., Ribeiro-Dasilva, M. C., Rahim-Williams, B. & Riley, J. L. 3rd Sex, gender, and pain: a review of recent clinical and experimental findings. J. Pain 10, 447–485 (2009).
P., Mettina, M. F., Roman, & G., Fehm-Wolfsdorf Personality and olfactory sensitivity. J. Res. Pers. 32, 510–518 (1998).
Eysenck, H. J. & Eysenck, M. W. Personality and Individual Differences (Plenum Press: New York, NY, USA (1985).
Johnson, K. G. & Van Vactor, D. Receptor protein tyrosine phosphatases in nervous system development. Physiol. Rev. 83, 1–24 (2003).
Van Vactor, D. Protein tyrosine phosphatases in the developing nervous system. Curr. Opin. Cell Biol. 10, 174–181 (1998).
De Fruyt, F., Van De Wiele, L. & Van Heeringen, C. Cloninger's psychobiological model of temperament and character and the five-factor model of personality. Pers. Individ. Dif. 29, 441–452 (2000).
Zhang, P., Xiang, N., Chen, Y., Sliwerska, E., McInnis, M. G., Burmeister, M. et al. Family-based association analysis to finemap bipolar linkage peak on chromosome 8q24 using 2500 genotyped SNPs and 15 000 imputed SNPs. Bipolar. Disord. 12, 786–792 (2010).
Chen, C., Zhou, Z., Li, M., Qu, M., Ma, Q., Zhong, M. et al. Presenilin-2 polymorphisms and risk of sporadic AD: evidence from a meta-analysis. Gene 503, 194–199 (2012).
Shamir, A., Kwon, O. B., Karavanova, I., Vullhorst, D., Leiva-Salcedo, E., Janssen, M. J. et al. The importance of the NRG-1/ErbB4 pathway for synaptic plasticity and behaviors associated with psychiatric disorders. J. Neurosci. 32, 2988–2997 (2012).
Rutten, K., Van Donkelaar, E. L., Ferrington, L., Blokland, A., Bollen, E., Steinbusch, H. W. et al. Phosphodiesterase inhibitors enhance object memory independent of cerebral blood flow and glucose utilization in rats. Neuropsychopharmacology 34, 1914–1925 (2009).
Tang, J., Chen, X., Xu, X., Wu, R., Zhao, J., Hu, Z. et al. Significant linkage and association between a functional (GT)n polymorphism in promoter of the N-methyl-D-aspartate receptor subunit gene (GRIN2A) and schizophrenia. Neurosci. Lett. 409, 80–82 (2006).
Wang, K. S., Liu, X. F. & Aragam, N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr. Res. 124, 192–199 (2010).
Fon, E. A., Sarrazin, J., Meunier, C., Alarcia, J., Shevell, M. I., Philippe, A. et al. Adenylosuccinate lyase (ADSL) and infantile autism: absence of previously reported point mutation. Am. J. Med. Genet. 60, 554–557 (1995).
Vogler, C., Spalek, K., Aerni, A., Demougin, P., Muller, A., Huynh, K. D. et al. CPEB3 is associated with human episodic memory. Front. Behav. Neurosci. 3, 4 (2009).
Denkova, E., Dolcos, S. & Dolcos, F. Reliving emotional personal memories: affective biases linked to personality and sex-related differences. Emotion 12, 515–528 (2012).
Carlson, R. & Levy, N. Studies of Jungian typology. I. Memory, social perception, and social action. J. Pers. 41, 559–576 (1973).
Cuttler, C. & Graf, P. Personality predicts prospective memory task performance: an adult lifespan study. Scand. J. Psychol. 48, 215–231 (2007).
Prinzie, P., Stams, G. J., Dekovic, M., Reijntjes, A. H. & Belsky, J. The relations between parents' Big Five personality factors and parenting: a meta-analytic review. J. Pers. Soc. Psychol. 97, 351–362 (2009).
Haan, N., Millsap, R. & Hartka, E. As time goes by: change and stability in personality over fifty years. Psychol. Aging. 1, 220–232 (1986).
Hogan, R. A Socioanalytic Perspective on the Five-Factor Model, (Guilford Press: New York, NY, USA, 1996).
Zhong, H. & Prentice, R. L. Correcting ‘winner’s curse’ in odds ratios from genomewide association findings for major complex human diseases. Genet. Epidemiol. 34, 78–91 (2010).
Acknowledgements
The genotype data of the discovery phase were gratefully made available by the Center for Genome Science, Korea National Institute of Health, Korea Centers for Disease Control and Prevention. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2010-0026606).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Kim, HN., Roh, SJ., Sung, Y. et al. Genome-wide association study of the five-factor model of personality in young Korean women. J Hum Genet 58, 667–674 (2013). https://doi.org/10.1038/jhg.2013.75
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2013.75