Abstract
Several lines of evidence have pointed out that genetic components have roles in thyrotoxic hypokalemic periodic paralysis (TTPP). In this study, for the first time we performed genome-wide association study (GWAS) in male hyperthyroid subjects in order to identify genetic loci conferring susceptibility to TTPP. We genotyped 78 Thai male TTPP cases and 74 Thai male hyperthyroid patients without hypokalemia as controls with Illumina Human-Hap610 Genotyping BeadChip. Among the SNPs analyzed in the GWAS, rs312729 at chromosome 17q revealed the lowest P-value for association (P=2.09 × 10−7). After fine mapping for linkage disequilibrium blocks surrounding the landmark SNP, we found a significant association of rs623011; located at 75 kb downstream of KCNJ2 on chromosome 17q, reached the GWAS significance after Bonferroni's adjustment (P=3.23 × 10−8, odds ratio (OR)=6.72; 95% confidence interval (CI)=3.11–14.5). The result was confirmed in an independent cohort of samples consisting of 28 TTPP patients and 48 controls using the same clinical criteria diagnosis (replication analysis P=3.44 × 10−5, OR=5.13; 95% CI=1.87–14.1; combined-analysis P=3.71 × 10−12, OR=5.47; 95% CI=3.04–9.83).
Similar content being viewed by others
Introduction
Thyrotoxic hypokalemic periodic paralysis (TTPP) is a rare complication of thyrotoxicosis characterized by episodes of intracellular shift of potassium ions and muscle weakness. It is reported mainly in young adult Asian males, occasionally reported in American Indians and Hispanics, but rarely in Caucasians and Africans. The clinical hallmarks resemble autosomal dominant hypokalemic periodic paralysis (HOKPP). Even though the underlying pathogenesis is not well understood, it has been defined as endocrine channelopathy. As this condition affects primarily males and people of Asian descent, underlying genetic susceptibility and high thyroid hormone levels may have roles in its pathogenesis. Candidate gene association study using markers of genes related to the biological pathway of TTPP have been performed; for examples, the human leukocyte antigen,1 Na+/K+-ATPase pump,2 the β2-adrenergic receptor gene3 and causative ion channel defects reported in HOKPP; that is, CACNA1S,4 SCN4A,5, 6 KCNE37 and KCNJ2.8 Mostly, neither mutation nor positive association was reported.9, 10, 11, 12 The failure of candidate gene association analyses is mainly due to the narrow range of target genes, as the basic pathogenesis of TTPP is unclear. Until recently Ryan et al.13 applied candidate gene approach by screening all the genes encoding potassium channels expressed in skeletal muscle with promoter containing thyroid hormone response elements. They discovered new potassium channel gene; KCNJ18, a gene encoding the inwardly rectifying potassium (Kir) channel, Kir2.6 on chromosome 17p11. The authors described five missense mutations (R205H, T354M, K366R, R399X and Q407X) and one frame shift mutation in TTPP patients. Notably, even with rationalized candidate gene approach, the mutations were demonstrated only in 33% of cases, which were 5 of 30 TTPP patients from Brazil, France and US, 1 of 83 individuals from Hong Kong and 7 of 27 from Singapore, but none from Thailand had the mutations. Their results pointed out the possible genetic heterogeneity underlying TTPP.
Recently, genome-wide association studies (GWAS) have been proven to be a powerful tool to identify susceptibility genes for common diseases, which are likely to enable us to reveal the roles of common polymorphisms on pathogenesis of disease.14 In this study, we attempted the first GWAS to identify genetic loci conferring susceptibility to TTPP, and here we described genetic locus near KCNJ2 that was associated with TTPP in Thai subjects.
Materials and methods
Subjects
All the subjects that participated in this study were unrelated Thai patients. The inclusion criteria of TTPP cases for both initial screening and later replication studies were hyperthyroid patients who presented with weakness as well as hypokalemia (K+ <3.5 mEq l−1) and low urine potassium concentration (spot urine K+ <15 mEq l−1), which indicated intracellular shift of potassium. Control subjects were recruited from hyperthyroid male patients with normal potassium levels during their hyperthyroid states. For an initial screening study, a total of 78 TTPP patients and 74 control subjects were recruited from three hospitals in Bangkok (Ramathibodi, King Chulalongkorn and Theptarin Hospital). The mean age of cases and controls were 40.5±2.0 (range 22–64) and 44.3±2.1 years (range 15–77), respectively (Supplementary Table 1). For the replication study, a second set of samples consisting of 28 TTPP patients and 48 controls were recruited from five Hospitals from different parts of Thailand (Ramathibodi, Maharat Nakhonratchasima Hospital, Prince of Songkla, Sappasitprasong and Ratchaburi Hospital). All cases and controls were male and their etiology of hyperthyroidism was Graves’ disease. This study was reviewed and approved by the Ethics Committee of Ramathibodi Hospital, Mahidol University and the Institutes of Physical and Chemical Research (RIKEN), Yokohama, Japan. All participants gave their written informed consent before participating in the study.
Genotyping and case–control association studies
A genome-wide analysis was conducted using Illumina Human-Hap610 Genotyping BeadChip according to the manufacturer's protocols (Illumina, INC., San Diego, CA, USA). We applied standard SNP quality control filters to exclude SNPs with low call rate (<99%), a Hardy–Weinberg equilibrium P<1.0 × 10−6 for controls. In the end, we obtained 498 465 autosomal SNPs and 9928 SNPs of X chromosome for the analysis. A principal component analysis (PCA) was performed via an ‘Eigen analysis’ in the computer program smartpca from the EIGENSOFT package15 to confirm the absence of significant genetic stratification between case and controls. Genotype data for the cases and controls and general population subjects for 89 East–Asian individuals (44 Japanese and 45 Han Chinese) from the International HapMap project16 were analyzed for the PCA. PCA plots were obtained using the first two components (Eigenvectors 1 and 2). To validate the genotyping results, we performed genotyping by means of multiplex PCR-based Invader assays (Third Wave Technologies, Madison, WI, USA)17 and compared the data obtained by the two platforms.
For fine mapping study, we plotted linkage disequilibrium blocks of pairwise D’ using Haploview software18 based on the HapMap phase 2 CHB population (http://www.hapmap.org/index.html.en). To further analyze SNPs within the linkage disequilibrium regions including the landmark SNP (rs312729), 26 haplotype-tagging SNPs (tag-SNPs; squared correlation coefficient between the two SNPs (r2) >0.8, minor allele frequency >0.05) were selected from the HapMap database and genotyped on 77 cases and 72 controls by multiplex PCR-based invader assay.
Statistical analyses
For association studies, the allele and genotype distributions in cases and controls were compared and evaluated in allelic, dominant- and recessive-inheritance models by two-tail Fisher's exact test. In the GWAS, SNPs were sorted according to the lowest P-value in one of these models. Significance levels were 9.83 × 10−8 (0.05/508 393) and 0.05 in the GWAS and the replication study, respectively. Odds ratios (OR) and confidence intervals (CI) were calculated using the non-risk genotype (12+22) as reference.
Results
Case–control association studies
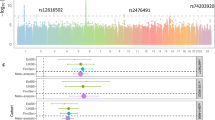
We first genotyped all cases and controls with Illumina Human-Hap610 Genotyping BeadChip. After excluding one case and two controls that were judged to be the outliers in the PCA (Supplementary Figure 1), we applied SNP quality control (call rate of 0.99 in both cases and controls). Of 600 420 SNPs genotyped, 498 465 autosomal SNPs and 9928 SNPs of X chromosome passed the quality control and were further analyzed. The genomic control inflation factor (GC) was 1.085, indicating a low possibility of false-positive associations resulting from population stratification (Supplementary Figure 2). Among the SNPs analyzed in the GWAS, although no SNPs showed genome-wide significant association with TTPP after the correction of multiple testing, rs312729 at chromosome 17 revealed the lowest P-value by allelic model for association (P=2.09 × 10−7, Figure 1, Supplementary Table 2).
Log10 P-value plots from a genome-wide association study (GWAS). Each dot represents P-value obtained from the GWAS using 77 TTPP cases and 72 controls. The Y-axis represents the −log10 of the minimal P-values calculated by Fisher's exact tests for three models: dominant, recessive and allelic model.
To validate the genotyping results from the Illumina assay, we re-genotyped all subjects for top 20 SNPs showing the smallest P-values in the first-stage GWAS and found the genotypes were concordant between the two platforms (data not shown).
Fine mapping
To further define a genomic region of interest, we genotyped 26 tag-SNPs in two linkage disequilibrium blocks surrounding the landmark SNP rs312729 (Supplementary Table 3) and 31 additional SNPs captured by those 26 tag-SNPs showing the minimal P-value <0.01 identified from the GWAS (Supplementary Table 4). We found rs623011 as the most significantly associated with the TTPP after Bonferroni's correction (P=3.23 × 10−8 by allelic model, OR=6.72; 95% CI=3.11–14.5) (Table 1). The rs623011 locates on chromosome 17q at 75 kb downstream of KCNJ2 gene (Figure 2).
Log10 P-value plots, linkage disequilibrium (LD) map and genomic structure of the region in chromosome 17q24.3 near KCNJ2. Fine mapping was performed in the region from 65.65 to 65.85 Mb. Black and red dots represent log10 P-value obtained from the GWAS and fine mapping using GWAS samples, respectively. Pairwise r2 was based on the genotype data of the 77 TTPP cases and 72 controls. An SNP rs623011 that is 75 kb downstream to KCNJ2 shows the most significant association with TTPP. MAF, minor allele frequency.
Replication study
After the fine mapping, we further evaluated the rs623011 in an independent set of samples consisting of 28 TTPP patients and 48 controls by PCR-based Invader assay. The association of rs623011 was replicated (P=3.44 × 10−5, OR=5.13; 95% CI=1.87–14.1; combined-analysis P=3.71 × 10−12, OR=5.47; 95% CI=3.04–9.83) (Table 1).
Discussion
In this study, we were able to demonstrate a new susceptible locus in TTPP patients using the GWAS approach. Currently there are two susceptible genetic loci associated with TTPP reported in the OMIM database, which are TTPP1 (OMIM: 188580) conferred by polymorphism in the CACNA1S gene on chromosome 1q322 and TTPP2 (OMIM: 613239) conferred by variation in the KCNJ18 gene on chromosome 17p11.13 We performed an association study by genotyping tag-SNPs surrounding CACNA1S, and direct sequencing of KCNJ18 in all TTPP cases, but we could not replicate the previously-published findings (data not shown).
The frequencies of a disease allele A of SNP rs623011 were lower in HapMap-CEU, compared with combined HapMap-CHB and HapMap-JPT (0.26 vs 0.48 respectively), and the frequency was highest in our TTPP cases (the frequency of the A allele=0.77 from the combined population). High frequencies of the A allele of SNP rs623011 in Asians may reflect the epidemiological finding of the higher TTPP prevalence in Asian populations, although the biological significance of how rs623011 contributes to the TTPP susceptibility should be elucidated.
The most significantly associated marker, rs623011 was located on chromosome 17q24.3 at 75 kb downstream to KCNJ2. The associations between rs623011, rs312729 and KCNJ2 expression quantitative trait locus (eQTL) were further analyzed using SNPEXP v1.119 (http://app3.titan.uio.no/biotools/tool.php?app=snpexp). The SNP data were derived from HapMap database and the expression level of KCNJ2 were based on GENEVAR20 (http://www.sanger.ac.uk/resources/software/genevar/), a database of gene expression from EBV-transformed lymphoblastoid cell lines. Only 210 unrelated samples from 270 available HapMap samples were included in this analysis. Unfortunately rs623011 was not successfully genotyped in HapMap samples; therefore eQTL analysis was carried out by Kruskal–Wallis test in R for the normalized expression of KCNJ2 against the genotype status (AA, AG, GG) of rs312729 only. A significant difference in normalized expression levels of KCNJ2 among genotypes of rs312729 was found (P=0.0329, Supplementary Figure 3). As rs623011 and rs312729 are in complete linkage disequilibrium (r2=0.942 in Hapmap phase 3 data), similar functional effects of genotypes on the KCNJ2 expression should be observed.
KCNJ2 encodes the Inward-rectifying K+ channel Kir2.1,21 which is highly expressed in cardiac and skeletal muscles. Mutations of KCNJ2 cause an autosomal dominant Andersen–Tawil syndrome8 (also known as Andersen syndrome: OMIM: 170390) that consists of cardiac arrhythmia, periodic paralysis and dysmorphic face. We performed direct sequencing of KCNJ2 but we could not find any deleterious mutations in our TTPP cases (data not shown). As previously described, transcriptional regulatory elements of genes can be located at regions distant from the transcription units.22 The risk allele A of rs623011, which is present at 75 kb downstream to KCNJ2, may affect the transcription of the gene and result in periodic paralysis similar to one of the clinical hallmarks in Andersen–Tawil syndrome patients.
In summary, we demonstrate that the genetic variant rs623011 is a new susceptible locus for TTPP. Our finding is valid for males TTPP and has to await further replication in women. Additional studies on other ethnic populations will also provide detailed information on the genetic etiology and heterogeneity of TTPP.
References
Yeo, P. P., Chan, S. H., Lui, K. F., Wee, G. B., Lim, P. & Cheah, J. S. HLA and thyrotoxic periodic paralysis. Br. Med. J. 2, 930 (1978).
Kung, A. W., Lau, K. S., Fong, G. C. & Chan, V. Association of novel single nucleotide polymorphisms in the calcium channel alpha 1 subunit gene (Ca(v)1.1) and thyrotoxic periodic paralysis. J. Clin. Endocrinol. Metab. 89, 1340–1345 (2004).
Kim, T. Y., Song, J. Y., Kim, W. B. & Shong, Y. K. Arg16Gly polymorphism in beta2-adrenergic receptor gene is not associated with thyrotoxic periodic paralysis in Korean male patients with Graves’ disease. Clin. Endocrinol. (Oxf). 62, 585–589 (2005).
Fontaine, B., Vale-Santos, J., Jurkat-Rott, K., Reboul, J., Plassart, E., Rime, C. S. et al. Mapping of the hypokalaemic periodic paralysis (HypoPP) locus to chromosome 1q31-32 in three European families. Nat. Genet. 6, 267–272 (1994).
Bulman, D. E., Scoggan, K. A., van Oene, M. D., Nicolle, M. W., Hahn, A. F., Tollar, L. L. et al. A novel sodium channel mutation in a family with hypokalemic periodic paralysis. Neurology 53, 1932–1936 (1999).
Jurkat-Rott, K., Mitrovic, N., Hang, C., Kouzmekine, A., Iaizzo, P., Herzog, J. et al. Voltage-sensor sodium channel mutations cause hypokalemic periodic paralysis type 2 by enhanced inactivation and reduced current. Proc. Natl Acad. Sci. USA 97, 9549–9554 (2000).
Abbott, G. W., Butler, M. H., Bendahhou, S., Dalakas, M. C., Ptacek, L. J. & Goldstein, S. A. MiRP2 forms potassium channels in skeletal muscle with Kv3.4 and is associated with periodic paralysis. Cell 104, 217–231 (2001).
Plaster, N. M., Tawil, R., Tristani-Firouzi, M., Canun, S., Bendahhou, S., Tsunoda, A. et al. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen's syndrome. Cell 105, 511–519 (2001).
Chen, L., Lang, D., Ran, X. W., Joncourt, F., Gallati, S. & Burgunder, J. M. Clinical and molecular analysis of Chinese patients with thyrotoxic periodic paralysis. Eur. Neurol. 49, 227–230 (2003).
Tang, N. L., Chow, C. C., Ko, G. T., Tai, M. H., Kwok, R., Yao, X. Q. et al. No mutation in the KCNE3 potassium channel gene in Chinese thyrotoxic hypokalaemic periodic paralysis patients. Clin. Endocrinol. 61, 109–112 (2004).
Schalin-Jantti, C., Laine, T., Valli-Jaakola, K., Lonnqvist, T., Kontula, K. & Valimaki, M. J. Manifestation, management and molecular analysis of candidate genes in two rare cases of thyrotoxic hypokalemic periodic paralysis. Horm. Res. 63, 139–144 (2005).
Wang, W., Jiang, L., Ye, L., Zhu, N., Su, T., Guan, L. et al. Mutation screening in Chinese hypokalemic periodic paralysis patients. Mol. Genet. Metab. 87, 359–363 (2006).
Ryan, D. P., da Silva, M. R., Soong, T. W., Fontaine, B., Donaldson, M. R., Kung, A. W. et al. Mutations in potassium channel Kir2.6 cause susceptibility to thyrotoxic hypokalemic periodic paralysis. Cell 140, 88–98 (2010).
Kruglyak, L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 22, 139–144 (1999).
Patterson, N., Price, A. L. & Reich, D. Population structure and Eigen analysis. PLoS Genet. 2, e190 (2006).
International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
Ohnishi, Y., Tanaka, T., Ozaki, K., Yamada, R., Suzuki, H. & Nakamura, Y. A high-throughput SNP typing system for genome-wide association studies. J. Hum. Genet. 46, 471–477 (2001).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Holm, K., Melum, E., Franke, A. & Karlsen, T. H. SNPexp - a web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinformatics 11, 600 (2010).
Yang, T. P., Beazley, C., Montgomery, S. B., Dimas, A. S., Gutierrez-Arcelus, M., Stranger, B. E. et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 26, 2474–2476 (2010).
Derst, C., Karschin, C., Wischmeyer, E., Hirsch, J. R., Preisig-Muller, R., Rajan, S. et al. Genetic and functional linkage of Kir5.1 and Kir2.1 channel subunits. FEBS Lett. 491, 305–311 (2001).
Bartkuhn, M. & Renkawitz, R. Long range chromatin interactions involved in gene regulation. Biochim. Biophys. Acta. 1783, 2161–2166 (2008).
Acknowledgements
The work described in this manuscript was supported by grants from the Thailand Research Fund RMU5080059 and the DMSc-RIKEN collaboration for genotyping support to researchers in Thailand.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Jongjaroenprasert, W., Phusantisampan, T., Mahasirimongkol, S. et al. A genome-wide association study identifies novel susceptibility genetic variation for thyrotoxic hypokalemic periodic paralysis. J Hum Genet 57, 301–304 (2012). https://doi.org/10.1038/jhg.2012.20
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.20
Keywords
This article is cited by
-
The clinical and genetic features in a cohort of mainland Chinese patients with thyrotoxic periodic paralysis
BMC Neurology (2015)
-
Inward-rectifying potassium channelopathies: new insights into disorders of sodium and potassium homeostasis
Pediatric Nephrology (2015)
-
Severe exacerbation of Andersen–Tawil syndrome secondary to thyrotoxicosis
Journal of Human Genetics (2014)
-
Genome-wide association study identifies a susceptibility locus for thyrotoxic periodic paralysis at 17q24.3
Nature Genetics (2012)
-
Thyrotoxic periodic paralysis: clinical and molecular aspects
Endocrine (2012)