Abstract
The relationship between Y-chromosome single-nucleotide polymorphisms (SNPs) registered in the Japanese SNP (JSNP) database (http://snp.ims.u-tokyo.ac.jp) and Y-binary haplogroup lineages was investigated to identify new Y-chromosomal binary haplogroup markers and further refine Y-chromosomal haplogroup classification in the Japanese population. We used SNPs for which it was possible to construct primers to make Y-specific PCR product sizes small enough to obtain amplification products even from degraded DNA, as this would allow their use not only in genetic but also in archeological and forensic studies. The genotype of 35 JSNP markers were determined, of which 14 were assigned to appropriate positions on the Y-chromosomal haplogroup tree, together with 5 additional new non-JSNP markers. These markers defined 14 new branches (C3/64562+13, C3/2613-27, D2a1b/006841*, D2a1b/119166-11A, D2a/022456*, D2a/119166-11A, D2a/119167rec/119167-40rec*, D2a/75888-GC, O3a3c/075888-9T/10T*, O3a3c/075888-9T/9T, O3a3/8425+6, O3a3/119166-13A*, O3a3/008002 and O3a4/037852) and 21 new internal markers on the 2008 Y-chromosome haplogroup tree. These results will provide useful information for Y-chromosomal polymorphic studies of East Asian populations, particularly those in and around Japan, in the fields of anthropology, genetics and forensics.
Similar content being viewed by others
Introduction
Y-chromosome polymorphisms are of particular interest in human evolutionary studies and forensic and medical genetics.1, 2, 3, 4, 5, 6, 7 Two types of Y-chromosome polymorphisms have been studied: Y short tandem repeats (Y-STRs) and Y-binary markers. Y-STR polymorphisms are widely used in the field of forensic medicine because of their higher level of diversity than that of Y-binary markers, and an enormous amount of Y-STR haplotype data has been accumulated (Y-STR haplotype reference database, www.yhrd.org). In contrast, Y-binary polymorphisms have mainly attracted interest in anthropological and evolutionary studies. In 2002, the Y-chromosome consortium (YCC) constructed a highly resolved tree of 153 binary haplogroups by genotyping 243 binary markers on a common set of samples.5 This tree was later modified.6 Using this modified tree, many researchers have identified new markers by comparing individual mutations. The tree was again updated and revised in 2008 with the inclusion of 600 binary markers and subdivision into 311 distinct haplogroups.7 Increasing the number of the bottom-most Y-binary markers deepens our understanding of recent movements and expansion of the human population, which is of interest from an anthropological perspective. It also provides a better means of personal identification in the forensic field. Increasing the number of defining or old internal markers may provide a clue to understanding ancient human expansion and movement through a comparison with known equivalent markers in the Y-chromosomal haplogroup tree. In addition, samples indicating paralogous sequence differences could help our understanding of gene-conversion events among such sequences on the human Y chromosome.8, 9, 10 Therefore, markers on the non-recombining portions of the Y chromosome offer huge potential in their application to anthropological, evolutionary and forensic studies.
Several studies have investigated peopling of Japan using Y-chromosomal binary markers.11, 12, 13, 14 However, these studies have mainly only used information on major haplogroups, and investigations based on more detailed haplogroup classification remain to be made because of the lack of suitable markers. Our group has already described the most detailed Y-chromosomal haplogroup classification in the Japanese population by expanding the YCC tree of 2003, assigning five new Japanese single-nucleotide polymorphism (JSNP) markers.15 These markers have now been included in the YCC tree of 2008.7 In the present study, we further searched for new Y-chromosomal binary haplogroup markers in the JSNP database16, 17 to classify Japanese Y-chromosomal haplogroups in greater detail. However, we adopted JSNPs for which it was possible to construct primers to make Y-specific PCR product sizes small enough to obtain amplification products even from degraded DNA, as this would allow their use not only in genetic but also in archeological and forensic studies.
Materials and methods
Samples
Genomic DNA was extracted from blood samples from healthy unrelated Japanese male individuals with informed consent. Genomic DNA of Malay individuals was extracted from tooth samples living in and around Kuala Lumpur. Appropriate consent was obtained from the patients. This study was approved by the ethics committee of Tokyo Dental College (approval no. 202 and 204) and met the conditions for cooperative study at the University of Malaya. The main set of Japanese samples included 263 individuals identical to those in an earlier study,15 in which we further detailed haplogroups at the level of the 2003 YCC tree, together with 16 Y-STR polymorphisms. If new mutations were found in a haplogroup, the number of samples from the Japanese population (78 samples) was increased in addition to the main set of samples.
Target for analysis and detection of biallelic markers
At the time of revision on 4 April 2012, 126 SNPs on the Y chromosome were registered in the JSNP database, of which 5 had already been assigned to the Y-chromosomal haplogroup tree.7, 15 Therefore, in this study, we analyzed information on the remaining 121 SNPs. According to the description in JSNP database, a total of 56 were located at pseudo-autosomal regions, 32 carried a single copy, 28 had multiple counterparts and 7 were uncertain for the chromosomal position.
Amplification by PCR was performed in a 40-μl mixture containing 10 ng genomic DNA, 10 mM Tris-HCl at pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.02% gelatin, 200 μM dNTP, 800 nM each primer and 1.75U AmpliTaq Gold (Applied Biosystems). The PCR primer sequences, amplification product sizes, mutation positions, mutation patterns, annealing temperatures and chromosomal positions for 35 biallelic markers taken from the JSNP database (http://snp.ims.u-tokyo.ac.jp)16, 17 and 5 markers newly found in this study are shown in Supplementary Table S1. A two-step PCR amplification process was used: 95 °C for 10 min, followed by 35 cycles of denaturation at 95 ° for 50 s and annealing and extension at an appropriate temperature for 105 s. After the 35th cycle, a final extension step was performed at the annealing and extension temperature for 10 min. Mutations were searched for by single-strand conformation polymorphism (SSCP) gel electrophoresis. An SSCP analysis was performed in a 17% polyacrylamide gel as described by Fujita and Kiyama,18 modified so that the gel contained 5% glycerin; the gel and reservoir buffer were 0.5 × TBE, and a 16 × 36-cm2 gel of 0.4 cm in thickness was used. Electrophoresis was performed at a constant voltage of 55 V cm−1 at 17.5 °C. All PCR products were visualized by silver staining. When differences were found in the SSCP gel electrophoresis, PCR products of different types were directly sequenced or reamplified from the bands on SSCP gels and sequenced. The nucleotide sequence of all types of sample, with or without mutations, was confirmed by sequencing.
Sequence analysis
The BigDye Terminator v1.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Austin, TX, USA) was used for PCR. Excessive dye was removed using Performa DTR gel filtration cartridges (EdgeBio, www.edgebio.com). Sequence analysis was performed on an ABI 3130 DNA Sequencer (HItachi HIgh-Technologies Corporation, Tokyo, Japan).
Construction of primers and determination of polymorphisms
Our goal was to search for haplogroup markers in the JSNP database for which we could construct primers to make Y-specific PCR product sizes, small enough to obtain amplification products from degraded DNA. Therefore, we designed PCR products smaller than approximately 200 bp. We first searched for sequences similar to those surrounding the mutation position and tried to construct primers specific to the target sequences, except where they fell into pseudo-autosomal regions. Female DNA was always used in PCR amplification as a control to determine whether the amplification products were obtained from the X chromosomes or not. When nontarget faint bands were obtained in the PCR amplification, the annealing temperature was increased to diminish these products. If same-sized PCR products originating in chromosomes other than the Y chromosome were amplified simultaneously, they were compared by SSCP gel electrophoresis and confirmed by sequencing to determine whether Y-specific bands could be discriminated from other PCR products. Some of the Y-chromosomal loci could not be discriminated from their counterparts on the X chromosome because they shared the same sequence. In these cases, we compared samples derived from different haplogroups by SSCP analysis, and if no difference was observed, we judged that there were no polymorphic differences on either the X or Y chromosomes.
Process of sample selection for determining new binary markers
The following procedure was basically applied to detect new binary markers in the JSNP database:
(1) We have already classified Y-binary haplogroups in 263 Japanese male individuals in 20 haplogroups (Supplementary Table S2).15 These samples were used as the basic set of samples in this study. The number of haplogroups in our original report15 decreased from 20 to 18 because the LINE1 marker was removed from the YCH Tree 2008. However, a sample from LINE1+ individuals was included in the first search for new binary markers. In the first screening, a typical sample was selected from each of the 20 haplogroups, together with an additional sample in the O2b* haplogroup (KN152), as its combination of Y-STR haplotypes was somewhat different from other data on the O2b* haplogroup (Supplementary Table S2). Finally, 21 samples for the first screening included one sample each from C1, C3*, D1, D2a, D2b*, D2b1/M125, D2b1/022457, N/O, O1*, O2a*, O2b*, O2b* (rare type), O2b1, O3*, O3c* (LINE1+), O3/002611, O3/LINE1 del (LINE1−), O3/021354*, O3e*, O3e1* and Q1 haplogroup according to the YCC 2003 classification. These were typed for the JSNP markers in question.
(2) When a mutation was found in a certain haplogroup, all samples that belonged to the corresponding haplogroup were typed for the marker to investigate whether it defined the same corresponding haplogroup marker or subdivided the branch. In such cases, we further increased the number of samples in addition to the basic set of samples (Supplementary Table S2).
(3) When no mutation was found, 29 different samples, including those carrying different Y-STR haplotype characteristics within the haplogroups, were further selected from the basic set and typed for JSNP markers (Supplementary Table S2). Where no further mutations were found, this was subsequently regarded as no detection of that mutation in the present study.
Nomenclature of haplogroups and other remarks
To determine binary haplogroups, we referred to the Y-chromosomal haplogroup tree 2008 (the YCH Tree 2008).7 To clarify descriptions, we have used + and − for the presence and absence of mutations, respectively. In addition, in designating new lineages, we have not changed the original binary haplogroup name used in the YCH Tree 2008, so as not to confuse the present status of the tree. Therefore, the nomenclature for the new binary haplogroups is tentative. Although the JSNP ID is designated by the letters IMS-JST followed by a six-digit figure, we did not always use this symbol to shorten description. When a new mutation was found near the location of an IMS-JST SNP, it was designated by the original JSNP number and the distance in base pairs from the position of that IMS-JST SNP followed by + or − depending on whether it was downstream or upstream, respectively. In addition, when the number of the IMS-JST SNP started with a 0-- or 00--, the 0 was deleted from the description of the new mutation.
The Y chromosome contains many paralogous sequences.9 JSNP markers that have more than two copies may belong to this category. When primers were constructed, we searched for the same sequence as that of the PCR region by BLAT to identify the number of copies on the Y chromosome. These markers were also investigated, and several markers showed variation. In these cases, they usually displayed two types of PCR products in the SSCP gel. Each type was distinguished and expressed by a slash. Markers that had more than two copies in the BLAT search are referred to as multi-copy sequences in this article.
Results
Evaluation of 126 JSNPs as Y-chromosomal binary haplogroup markers
We investigated possibility of utilizing Y-chromosomal JSNPs as binary haplogroup markers. The results of our evaluation of 126 JSNPs on the Y chromosome are shown in Table 1. Among 126 JSNPs, the positions of 5 markers15 had already been assigned to the YCH Tree 2008 (Table 1, row 1). Among the remaining 121 markers, 14 were polymorphic and assigned in this study, including 4 multi-copied markers (Table 1, row 2). At 19 loci that included seven multi-copied markers, Y-specific products could be discriminated, but no mutation was found (Table 1, row 3; Supplementary Table S1). At 2 loci, X- and Y-specific products could not be discriminated in the SSCP gels, although they showed polymorphic differences (Table 1, row 4).
Evaluation of the remaining 86 markers was as follows. Although we examined polymorphisms at the two loci IMS-JST021351 and IMS-JST021352 (Table 1, row 5; Supplementary Table S1) in pseudo-autosomal regions, the results of typing were not useful for Y-chromosomal haplogroup analysis. Therefore, we did not further study the polymorphisms of the remaining 54 markers in pseudo-autosomal regions (Table 1, row 5). Among 30 multi-copied markers in JSNPs on the Y chromosome, 11 were described above. Suitable primers for 2 of the remaining 19 multi-copied markers could not be prepared because of sequence similarity with other counterparts on the Y chromosome (Table 1, row 10). We examined five multi-copied markers. However, the results were so complex that we have yet to understand the process of mutation, and so have omitted any discussion of them in this paper (Table 1, row 6). We have not yet examined the additional 12 other multi-copied markers for assignment to the YCH tree (Table 1, row 6). We further excluded 11 loci from the analysis because of the reasons given in Table 1 (row 7–9, 11).
Mutations in clade C
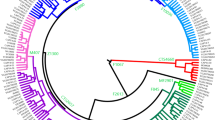
IMS-JST029149 carried a single copy on the Y chromosome, and mutations were found in the C1 and C3 lineages (Supplementary Table S2), suggesting that it was the defining marker of haplogroup C. We found three examples of Malay individuals belonging to haplogroup C carrying M216 T(+), but also RPS4Y711 C(−) (unpublished observation) (Supplementary Table S2). This indicates that the M216 mutation is older than the RPS4Y771 mutation. Because 029149 was T(+) in these samples, it was assigned to the same position as the defining marker, M216 (Figure 1a).
Y-chromosomal phylogenetic tree: (a) C and D clades and (b) O clade. Number of samples and frequencies of haplogroups are also indicated. New markers and lineages found in this study are shown in bold letters, and haplogroups and markers not examined are shown in light letters. In naming lineage, original states based on YCC 2003 in previous report15 are shown in parentheses.
IMS-JST037816 carried two copies on the Y chromosome, and mutations were found only in the C1 lineage (Supplementary Table S2; Figure 1a). Samples of C1 carried two bands corresponding to C(−) and T(+) in an SSCP gel, showing that the T(+) mutation at one of the loci of 037816 corresponded to the same position as the M105 mutation. When new samples were examined for 037816, the locus was not amplified in one sample (Supplementary Table S2, sample no. 1). Large-sized PCR amplification (214 bp) including original primer sequences (Supplementary Table S1) also failed to amplify the target, suggesting that failure of amplification was not caused by primer mismatch. This sample also lacked the Y-STR locus DYS438 (not shown). DYS438 was located in the middle of the two copies of 037816, whose chromosomal positions on the Y chromosome were 24287112 and 24591604, demonstrating that the sample had a defect of more than 300 kbp in size, including two 037816 loci.
When three samples of Malay individuals in the C lineage were amplified by large-sized 037816 primer pairs, a different type of SSCP pattern was obtained in one of them (Supplementary Table S2), which carried an A/G mutation at the 80th position upstream from the 037816 mutation (37816-80) (Supplementary Table S2). This mutation created a new branch between the M216 and RPS4Y711 mutations in the YCH tree (Figure 1a). Because this study focused on Japanese Y-chromosomal lineages, we have not included this branch in the Y-binary haplogroup tree as yet (Figure 1a).
IMS-JST064562 carried a single copy on the Y chromosome, and one sample in the C3 haplogroup had a mutation at the13th position downstream from 064562 (Supplementary Table S2). We designated this lineage C3/64562+13 (Figure 1a). This sample had a different and characteristic Y-STR haplotype in the C3* samples (repeat structure of DYS390 and combination of DYS385), suggesting that it belonged to a different haplogroup.
IMS-JST002612 and IMS-JST002613 were located adjacently and carried a single copy on the Y chromosome. Ten samples in the C3 haplogroup had a mutation at the 27th position upstream from the 002613 (Supplementary Table S2). Because the samples in this group had a similar type of Y-STR haplotype, we designated this lineage C3/2613-27 (Figure 1a). These samples carried a structural variation at the DYS438 locus, in which the repeat structure was (TTTTC)1(TTTTA)1(TTTTC)8, unlike the common (TTTTC)n structure.15, 19 The 2613-27 mutation was completely associated with the DYS438 structural variation in our samples. Examination of 9 additional samples in the C3* haplogroup also showed 2613-27(+) (Supplementary Table S2).
Mutation in clade D
IMS-JST008425 and 008426 carried a single copy each on the Y chromosome, and a mutation for each type was found in the D haplogroup (Supplementary Table S2). Because two samples of Malay individuals in the E haplogroup also carried these mutations, they were considered to correspond to the same position as the defining mutation, YAP (Figure 1a). Comparison with the list of YCC primers proved that the 008426 was identical to the M203.
IMS-JST055457 carried a single copy on the Y chromosome, and mutations were found in the D2a1b haplogroup and associated completely with the 022457 (Supplementary Table S2). Therefore, it was assigned to the same position as 022457 (Figure 1a).
IMS-JST006841 carried a single copy on the Y chromosome, and mutations were found in the D2a1b* haplogroup with similar distribution to the 022457 or 055457 mutation. However, when all of the D2a1b samples in the basic set of 263 individuals and 28 additional individuals were examined for 006841, one sample did not carry this mutation, indicating that this mutation occurred after 022457 and 055457 (Supplementary Table S2). We designated this lineage D2a1b/006841 (Figure 1a).
The chromosomal position of IMS-JST119166 carried a single copy on the Y chromosome. Although a similar sequence was found on chromosome 1, our primers did not amplify its counterpart on chromosome 1. A polymorphic region commonly included 12A repeats. However, two samples with an 11A repeat were found in one of the basic sets of the D2a1b/006841 haplogroup and in one of the 29 additional samples (Supplementary Table S2). Therefore, we designated this lineage D2a1b/119166-11A (Figure 1a). We propose that this lineage should be typed together with marker 006841 or 022457, in addition to the 119166-11A mutation.
IMS-JST022456 carried a single copy on the Y chromosome, and 17 of the 37 samples in D2a* had mutations and formed a new branch (Supplementary Table S2). We designated this lineage D2a/022456 (Figure 1a). One sample in this group had 11A repeats at the 119166 locus (Supplementary Table S2). We designated this lineage D2a/119166-11A. This lineage should also be typed together with marker 022456 (Figure 1a).
The mutation pattern of IMS-JST119167 was found to be a GTC deletion (GTCdel). 119167 carried four copies on the Y chromosome, and one of the counterparts obtained using BLAT search carried this mutation. There was another A/G mutation at the 40th position upstream from the 119167 mutation (119167-40) (rs2538946). The G mutations and GTCdels were completely associated in 115 samples examined in this study (Supplementary Table S2). Although the mutation pattern including G and GTCdel was basically found in all lineages, except the C haplogroup, two samples in the basic set and one in the additional samples in the D2a* haplogroup did not carry these mutations (Supplementary Table S2). We believe that recurrent mutation occurred in these samples. This mutation pattern was also found in one sample in the E haplogroup in the Malay population (Supplementary Table S2). Therefore, recurrent mutation may have occurred also in the Malay lineage. Because of the unique Y-STR haplotypes of the mutated samples in D2a* haplogroup of the Japanese population (Supplementary Table S2), we tentatively designated the lineage of these samples D2a/119167/119167-40rec* (Figure 1a). We propose that in determining this lineage, the M116 and 119167/119167-40 mutations must be confirmed simultaneously (Figure 1a).
IMS-JST075888 carried two copies on the Y chromosome. Although a polymorphic region of 075888 commonly included 10T repeats, one of the samples in the D2a/119167-40/119167rec haplogroup possessed a TT/GC mutation in the T repeat in one of the two copies (genotype GC+8T/10T) (Supplementary Table S2). Therefore, we designated this haplogroup D2a/075888-GC (Figure 1a). This mutation may have occurred very recently, because the STR haplotypes of samples SO47 and CB48 in the D2a/119167/119167-40rec* haplogroup were identical, suggesting close lineage between these samples.
Mutation in clade F
IMS-JST001552 and 003305 carried a single copy each on the Y chromosome. Both mutations were found in the N/O and O or Q haplogroups, respectively (Supplementary Table S2). Because these mutations were also found in the sample of the F* haplogroup in the Malay population, we assigned these markers to the defining position of the F clade (Figure 1a).
Mutation in clade O
IMF-JST075888 carried two copies on the Y chromosome, as pointed out above in the section on clade D. Another type of mutation was found in haplogroup O3a3c. Haplogroup O3a3c was classified into three types by different patterns of mutation: one possessed only a 10T repeat (10T/10T type); another possessed a 9T repeat in addition to the 10T repeat (9T/10T type); and the other possessed the 9T repeat only (9T/9T type) (Supplementary Table S2). Among 339 samples examined in this study, the 9T repeat was found only in this haplogroup, suggesting that mutation of the number of the T-stretch at 075888 is not a common phenomenon. We estimated the order of mutation as follows. The 9T repeat occurred in one of the repeats of 075888, producing the 9T/10T type. Next, the 10T repeat in this group further mutated into 9T, producing the 9T/9T type. Although mutation in the number of T-repeats is not a common phenomenon, we propose that these lineages must be typed together with O3a3c marker M134 to define the haplogroup name. Finally, we designated each haplogroup O3a3c/075888-9T/10T* and O3a3c/075888-9T/9T (Figure 1b).
One of the samples in the O3a3* haplogroup had a mutation at the 6th position downstream from IMF-JST008425 (Supplementary Table S2). 008425 carried a single copy on the Y chromosome. Therefore, we designated this haplogroup O3a3*/8425+6 (Figure 1b).
The 119166 locus carried a single copy on the Y chromosome, as pointed out in the section on clade D. Two samples in the O3a3* haplogroup had different types of mutation at 119166 (Supplementary Table S2). They carried 13A, which was different from the mutation type in the D2a1b lineage (11A). The Y-STR types of DYS393, DYS385 and DYS434 in these samples were unique in the O3a3* samples (Supplementary Table S2), suggesting that these samples belonged to different lineages in this haplogroup. The same type was further found in a new sample, and we designated this lineage O3a3/119166-13A* (Figure 1b). This lineage must be typed together with 021354 (=P201). Among these, one sample further possessed a mutation at locus IMS-JST008002, which carried a single copy on the Y chromosome (Supplementary Table S2). Therefore, we designated this lineage O3a3/008002 as a branch of the O3a3/119166-13 A* lineage (Figure 1b).
IMS-JST037852 carried two copies on the Y chromosome. Samples with G/C mutations in one of the two copies were found in the O3a4* haplogroup (Supplementary Table S2). This lineage was designated as O3a4/037852 (Figure 1b).
Population study
The number of haplogroups in the basic set of 263 samples increased from 18 to 32 (Figures 1a and b). Haplogroup diversity for binary polymorphisms was calculated to be 86.2% for the 18 haplogroups and 87.5% for the 32 haplogroups.
Discussion
The relationship between Y-chromosomal SNPs in the JSNP database and Y-chromosomal haplogroups was investigated. Among 19 markers assigned in this study, 6 carried multiple copies on the Y chromosome. Four of them, 037816, 37816-80, 075888 and 037852, carried two copies and the other two, 119167-40 and 119167, carried four copies. 119167-40 and 119167, which were located on the RBMY2B gene exhibited the most complex pattern. Three out of the four loci of 119167-40 and 119167 in the human genome database were 119167-40A(−) and 119167 GTC. One similar counterpart of the Pan troglodytes shared a non-deleted pattern with human 119167. Therefore, it is possible that this non-deleted pattern is original and that the deleted pattern is a mutation. If so, all of the Japanese lineages (D, N, O and Q), except the C lineage, show mutations 119167-40A(−)/G(+) and 119167 GTC/GTCdel. The present results suggest that recurrent mutation occurred at the D clade in the Japanese population and at the E clade in the Malay population. It is possible that gene conversion occurred on the Y-chromosomal paralogous sequences 119167-40 and 119167,8, 10 and that the mutated sequences 119167-40G(+) and 119167 GTCdel were eliminated. We have already showed a close association between Y-binary haplogroups and Y-STR haplotypes.15 The Y-STR haplotypes of the two samples in D2a/119167/119167-40* were identical and unique in the D2a haplogroup. Allele 13 in the DYS437 locus was found only in these samples in this study (Supplementary Table S2). Therefore, we apply mutations of 119167 and 119167-40 as a defining marker only to this lineage, and propose a new bottom-most haplogroup lineage, D2a/119167/119167-40*.
We propose three JSNPs showing variation in the number of the same nucleotide as markers for new haplogroup lineages. These mutations might also be considered to represent differences in single-nucleotide repeat polymorphisms rather than SNPs. However, these markers were assigned from a number of reasons. This type of mutation was found as 119166-11A in the two lineages in the D clade, and as 119166-13A, 075888-9T or 075888-9T/9T in the O clade. Each type of mutation was found only in a limited haplogroup and was not frequently found in other haplogroups. As was seen at the 075888 locus, the individual belonging to the 075888 haplogroup in O3a3 possessed somewhat different characteristics of Y-STR haplotype depending on each haplogroup (such as the allele type of DYS439 and DYS385), further suggesting that it constitutes a different lineage in the O3a3 haplogroup (Supplementary Table S2). In addition, M91, a defining mutation in the A clade, shows variation in the number of T-repeats (9T→8T), and has been used as a binary marker in the YCH tree.7, 20 Therefore, we propose assignment of these mutations as haplogroup markers. However, we also propose that these mutations can only be assigned to an appropriate position on the YCH tree if the established adjacent upstream internal marker is positive. As far as the 075888-9T or 075888-10T markers are concerned, it is possible that they were developed by gene conversion, as 07588 has two copies on the Y chromosome. Therefore, it is also possible that 9T/10T back-mutates to 10T/10T in some cases, although the present results are insufficient to confirm this.
This study identified 2 new branches and 5 internal markers in the C clade, in addition to the 19 branches and 30 internal markers in the YCH Tree 2008. Hou et al.21 studied the repeat structure of DYS438 in the Chinese population and demonstrated the presence of (TTTTC)1(TTTTA)1(TTTTC)8 repeat structure. As the C3/002613-27 samples contained the same repeat structure at the DYS438 locus, it is possible that this lineage is also present in the Chinese population.
We identified 2 defining mutations in the DE clade and 6 new branches and 8 internal markers in the D clade, in addition to the 8 defining mutations in the DE clade and 15 branches and 23 internal markers in the D clade shown in the YCH Tree 2008. D lineages have commonly been found in central Asia (Tibet) and Japan.7 Because the lineage in Tibet is mainly D1 or D3a,22 it is probable that the present markers mainly subdivide lineages in the Japanese population.
The O clade is a major lineage in East Asia. In all, 33 kinds of haplogroup and 44 kinds of internal marker subdividing 30 haplogroups have been proposed in the YCH Tree 2008. We have identified six new branches and six new internal markers in the O clade. Among these, five branches were found in the O3a3 lineage, suggesting a high level of diversity in this lineage in East Asia.
Two markers, 001552 and 003305, were tentatively assigned to the position of F. Although we have not yet excluded the possibility of assigning these markers to the H2 or H* lineages in the H clade, it is possible that they do not belong to these haplogroups, as the H haplogroup has not yet been found in the Japanese. Their possible assignment to other macrohaplogroups has already been excluded.
As described above, the relationship between JSNP markers and Y-binary haplogroups was investigated. In this study, the D2a1b/006841 and O2b1 lineages accounted for 45% of the Japanese male lineages. However, no markers have been discovered that would further subdivide these lineages, suggesting that they represent a very homogeneous population. Binary markers provide useful information in the field of human genetic and anthropological studies. It is necessary to increase new binary markers, which are characteristic of a certain geographic area, in order to use these information more effectively. In the field of forensics, binary markers also have the advantage of obviating the problem of allele drop against degraded or low copy number materials, as long as detection of polymorphisms is conducted carefully and without problematic contamination. These results suggest the potential of Y-chromosomal binary polymorphisms in studies on Japanese and south-east Eurasian populations.
References
Mitchell, R. J. & Hammer, M. F. Human evolution and the Y chromosome. Curr. Opin. Genet. Dev. 6, 737–742 (1996).
Underhill, P. A., Shen, P., Lin, A. A., Jin, L., Passarino, G., Yang, W. H. et al. Y chromosome sequence variation and the history of human populations. Nat. Genet. 26, 358–361 (2000).
Jobling, M. A. & Tyler-Smith, C. New uses for new haplotypes the human Y chromosome disease and selection. Trends Genet. 16, 356–362 (2000).
Gill, P., Brenner, C., Brinkmann, B., Budowle, B., Carracedo, A., Jobling, M. A. et al. DNA commission of the International Society of Forensic Genetics: recommendations on forensic analysis using Y-chromosome STRs. Int. J. Legal Med. 114, 305–330 (2001).
The Y Chromosome Consortium. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 12, 339–348 (2002).
Jobling, M. A. & Tyler-Smith, C. The human Y chromosome: an evolutionary marker comes of age. Nat. Rev. Genet. 4, 598–612 (2003).
Karafet, T. M., Mendez, F. L., Meilerman, M. B., Underhill, P. A., Zegura, S. L. & Hammer, M. F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 18, 830–838 (2008).
Rozen, S., Skaletsky, H., Marszalek, J. D., Minx, P. J., Cordum, H. S., Waterston, R. H. et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003).
Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P. J., Cordum, H. S., Hillier, L., Brown, L. G. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003).
Adams, S. M., King, T. E., Bosch, E. & Jobling, M. A. The case of the unreliable SNP: recurrent back-mutation of Y-chromosomal marker P25 through gene conversion. Forensic Sci. Int. 159, 14–20 (2006).
Hammer, M. F. & Horai, S. Y chromosomal DNA Variation and the peopling of Japan. Am. J. Hum. Genet. 56, 951–962 (1995).
Tajima, A., Hayami, M., Tokunaga, K., Juji, T., Matsuo, M., Marzuki, S. et al. Genetic origins of the Ainu inferred from combined DNA analyses of maternal and paternal lineages. J. Hum. Genet. 49, 187–193 (2004).
Hammer, M. F., Karafet, T. M., Park, H., Omoto, K., Harihara, S., Stoneking, M. et al. Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J. Hum. Genet. 51, 47–58 (2006).
Matsukusa, H., Oota, H., Haneji, K., Toma, T., Kawamura, S. & Ishida, H. A genetic analysis of the Sakishima islanders reveals no relationship with Taiwan aborigines but shared ancestry with Ainu and main-island Japanese. Am. J. Phys. Anthropol. 142, 211–223 (2010).
Nonaka, I., Minaguchi, K. & Takezaki, N. Y-chromosomal binary haplogroups in the Japanese population and their relationship to 16 Y-STR polymorphisms. Ann. Hum. Genet. 71, 480–495 (2007).
Haga, H., Yamada, R., Ohnishi, Y., Nakamura, Y. & Tanaka, T. Gene-based SNP discovery as part of the Japanese Millennium Genome Project: identification of 190562 genetic variations in the human genome, single-nucleotide polymorphism. J. Hum. Genet. 47, 605–610 (2002).
Hirakawa, M., Tanaka, T., Hashimoto, Y., Kuroda, M., Takagi, T. & Nakamura, Y. JSNP: a database of common gene variations in the Japanese population. Nucleic Acids Res. 30, 158–162 (2002).
Fujita, T. & Kiyama, M. Identification of DNA polymorphism by asymmetric-PCR SSCP. Biotechniques 19, 532–534 (1995).
Gusmao, L., Alves, C. & Amorim, A. Molecular characteristics of four human Y-specific microsatellites (DYS434, DYD437, DYS438, DYS439) for population and forensic studies. Ann. Hum. Genet. 65, 285–291 (2001).
Underhill, P. A., Passarino, G., Lin, A. A., Shen, P., Mirazon, M., Foley, R. A. et al. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann. Hum. Genet. 65, 43–62 (2001).
Hou, Y. P., Zhang, J., Li, Y. B., Wu, J., Zhang, S. Z. & Prinz, M. Allele sequences of six new Y-STR loci and haplotypes in the Chinese Han population. Forensic Sci. Int. 15, 147–152 (2001).
Gayden, T., Cadenas, A. M., Regueiro, M., Singh, N. B., Zhivotovsky, L. A., Underhill, P. A. et al. The Himalayas as a directional barrier to gene flow. Am. J. Hum. Genet. 80, 884–894 (2007).
Acknowledgements
We would like to thank Associate Professor Jeremy Williams, Laboratory of International Dental Information, Tokyo Dental College, for editing this manuscript. Part of this study was supported by the Ministry of Education, Science, Sports and Culture, Grant-in-Aid for Scientific Research (C) (22592345).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Naitoh, S., Kasahara-Nonaka, I., Minaguchi, K. et al. Assignment of Y-chromosomal SNPs found in Japanese population to Y-chromosomal haplogroup tree. J Hum Genet 58, 195–201 (2013). https://doi.org/10.1038/jhg.2012.159
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.159
Keywords
This article is cited by
-
Y chromosome analysis for common surnames in the Japanese male population
Journal of Human Genetics (2021)
-
A commentary on assignment of Y-chromosomal SNPs found in Japanese population to Y-chromosomal haplogroup tree
Journal of Human Genetics (2013)