Abstract
Fatty acid-binding protein (FABP) gene family encode fatty acid-binding proteins and consist of at least 12 members, of which FABP7, 5 and 3 are expressed in the brain. We previously showed that FABP7 is associated with schizophrenia and bipolar disorder. Recently, genetic overlap between autism and schizophrenia has been reported. Therefore, in this study, we set out to examine the possible roles of brain-expressed FABPs in autism, focusing primarily on potentially functional polymorphisms (that is, missense polymorphisms). First, we resequenced the three genes using 285 autism samples. We identified 13 polymorphisms, of which 7 are novel. Of the novel single-nucleotide polymorphisms (SNPs), two are missense mutations, namely, 376G>C (Val126Leu) in FABP7 and 340G>C (Gly114Arg) in FABP5. Second, we tested for the genetic association of four missense SNPs with autism and schizophrenia, but failed to detect significant results. Finally, as a web-based algorithm predicts that the 8A>G (Asp3Gly; rs17848124) in FABP3 is ‘probably damaging’, we estimated the possible impact of this SNP, and found that the loss of charge and salt bridge, caused by the Asp3-to-Gly3, may affect stability of the FABP3 protein. Future searches for associated phenotypes with missense SNPs using larger samples are highly warranted.
Similar content being viewed by others
Introduction
Schizophrenia and autism are devastating psychiatric illnesses, with the former showing mainly adulthood onset and the latter childhood onset. Recent research proposes common genetic mechanisms between the two diseases.1 As an additional environmental factor, the role of fatty acids has attracted attention in the pathophysiology of both diseases.2 Our interest is in fatty acid-binding proteins (FABPs) as mediator molecules for linking fatty acids to brain function. FABPs constitute a gene family of at least 12 members.3, 4 Brain-expressed FABPs include FABP7, FABP5 and FABP3.5 We previously performed association studies between the above three FABP genes and schizophrenia and bipolar disorder, and detected modest associations between FABP7 and schizophrenia and bipolar disorder.6, 7
In this study, we set out to examine the possible genetic roles for FABP7, FABP5 and FABP3 in autism. First, we resequenced the three FABP genes using 285 autism samples. We then focused on the identified missense single-nucleotide polymorphisms (SNPs), undertaking a genetic evaluation of these SNPs in autism and schizophrenia because of the possible overlap in mechanisms between the two diseases. We also discussed the possible impact of a potentially important SNP in terms of its predicted structural and functional consequences.
Materials and methods
Subjects
A total of 285 autistic patients of Japanese descent (236 men, 49 women; aged between 3 and 32 years) were used for the resequencing analysis and association studies. The diagnosis of autism was made on the basis of the Autism Diagnostic Interview-Revised (ADI-R)8 criteria. The detected missense polymorphisms were analyzed in association studies using the above mentioned autistic subjects, 1060 schizophrenics (diagnosed according to DSM-IV) (503 men, 557 women; mean age 48.0±13.8 years) and 1060 controls who are free of mental disorders (503 men, 557 women; mean age 47.7±13.6 years) during brief interviews by psychiatrists. Because we did not administer structural or semi-structural instruments for the recruitment of control subjects, it may be difficult to completely exclude the contamination of subjects with psychiatric problems, including Asperger syndrome. The study was approved by the ethics committees of the RIKEN and Hamamatsu University.
Polymorphism screening of FABP genes and genotyping
Protein-coding regions and exon/intron boundaries of FABP7, FABP5 and FABP3 were screened for polymorphisms by direct sequencing of PCR products (Supplementary Figure 1). The primers used for amplification and PCR conditions are listed in Supplementary Table 1. Custom TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA, USA) were used to score the identified missense SNPs, along with ABI PRISM 7900 Sequence Detection System and SDS v2.3 software (Applied Biosystems). The insertion/deletion polymorphisms were scored by inspecting the sequencing electrophelograms.
Results and discussion
Identification of missense SNPs in the FABP genes and association studies
Our polymorphism screen detected 13 different variants in FABP7, FABP5 and FABP3: 7 are novel and 6 were previously reported (Table 1). With respect to novel variants, the 376G>C (Val126Leu; SNP ID ‘ss160853728’ was obtained from NCBI) in FABP7 and 340G>C (Gly114Arg; newly assigned as ‘ss160853732’) in FABP5 were missense SNPs (Table 1). The total number of missense SNPs identified in this study is four. We tested these four missense SNPs for association with autism and schizophrenia, because of the potentially functional importance of missense polymorphisms. As autism is more prevalent in males than in females, the association analysis of autism samples was done separately in males and in females. However, none of them showed significant P-values, although the two new missense SNPs (Val126Leu in FABP7 and Gly114Arg in FABP5) were not seen in over 1000 control samples (Table 2). When samples of both sexes were combined, the associations with autism were not significant at all the SNPs (data not shown). The low frequency of minor alleles from the examined SNPs, suggests that larger sample sizes are needed to exclude the possibility of type II errors, a limitation of the current study. The other caution of this study includes that age is not matched between autism and control samples.
The subject with FABP7 Val126/Leu126 genotype is an 8-year-old male, whose diagnosis is autistic disorder. The intellectual examination using the Wechsler Intelligence Scale for Children-III (WISC-III) showed the following scores: FIQ (full scale IQ)=96, VIQ (verbal IQ)=94 and PIQ (performance IQ)=100. Regarding the ADI-R scores, domain A (social interaction)=26, B (communication and language)=14, C (restricted and repetitive behaviors)=3 and D (onset of disorder)=3. His parents are mentally healthy, and the 126Leu allele was transmitted from his mother (Supplementary Figure 2). The subject with FABP5 Gly114/Arg1114 genotype is a 33-year-old male, whose diagnosis is PDDNOS (pervasive developmental disorder not otherwise specified including atypical autism). He does not have any family history of mental disorders. The Wechsler Adult Intelligence Scale III (WAIS-III) scores were FIQ=95, VIQ=94 and PIQ=97. The ADI-R scores were domain A=5, domain B=5, domain C=2 and domain D=0. The DNA samples of his parents were not available.
Possible impact of the missense SNPs on the structure and function of FABP proteins
A tool-website ‘PolyPhen’ can estimate the possible impact of an amino acid substitution on the structure and function of a protein (http://genetics.bwh.harvard.edu/pph/). According to this algorithm, only the Asp3Gly mutation in FABP3 is predicted to be ‘probably damaging’ among the four missense SNPs (Table 1). The possible impact of the Thr61Met mutation in FABP7 is discussed elsewhere.6
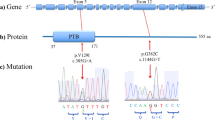
The crystal structure of FABP3 is known (Figure 1).9 Replacing Asp by Gly causes loss of a negative charge and breakage of a salt bridge between Asp3 and Lys45, suggesting that this missense polymorphism may affect protein stability. As FABP3 binds and transports fatty acids, destabilization of FABP3 could affect multiple biological processes.
The site of the missense polymorphism, Asp3Gly, in the FABP3 protein structure (PDB ID, 1HMT; complex with stearic acid).9 The salt bridge between Asp3 and Lys45 is shown as a dashed line, with its distance. This graphic was created using PyMOL (DeLano Scientific, Palo Alto, CA, USA).
References
Guilmatre, A., Dubourg, C., Mosca, A. L., Legallic, S., Goldenberg, A., Drouin-Garraud, V. et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch. Gen. Psychiatry 66, 947–956 (2009).
Kidd, P. M. Omega-3 DHA and EPA for cognition, behavior, and mood: clinical findings and structural-functional synergies with cell membrane phospholipids. Altern. Med. Rev. 12, 207–227 (2007).
Liu, R. Z., Li, X. & Godbout, R. A novel fatty acid-binding protein (FABP) gene resulting from tandem gene duplication in mammals: transcription in rat retina and testis. Genomics 92, 436–445 (2008).
Furuhashi, M. & Hotamisligil, G. S. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 7, 489–503 (2008).
Owada, Y. Fatty acid binding protein: localization and functional significance in the brain. Tohoku J. Exp. Med. 214, 213–220 (2008).
Watanabe, A., Toyota, T., Owada, Y., Hayashi, T., Iwayama, Y., Matsumata, M. et al. Fabp7 maps to a quantitative trait locus for a schizophrenia endophenotype. PLoS Biol. 5, e297 (2007).
Iwayama, Y., Hattori, E., Maekawa, M., Yamada, K., Toyota, T., Ohnishi, T. et al. Association analyses between brain-expressed fatty-acid binding protein (FABP) genes and schizophrenia and bipolar disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. (e-pub ahead of print 24 June 2009) (2009).
Lord, C., Rutter, M. & Le Couteur, A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 24, 659–685 (1994).
Young, A. C., Scapin, G., Kromminga, A., Patel, S. B., Veerkamp, J. H. & Sacchettini, J. C. Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids. Structure 2, 523–534 (1994).
Acknowledgements
We thank Drs Yamada and Hattori for help in recruiting samples, and the Research Resource Center of the RIKEN Brain Science Institute (BSI) for sequencing support. This work was supported by the RIKEN BSI Funds, CREST funds from the JST Agency, Grant-in-Aid from the MEXT and grants from the Mitsubishi Pharma Research Foundation. In addition, a part of this study is the result of ‘Development of biomarker candidates for social behavior’ study carried out under the Strategic Research Program for Brain Sciences by the MEXT of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
Rights and permissions
About this article
Cite this article
Maekawa, M., Iwayama, Y., Arai, R. et al. Polymorphism screening of brain-expressed FABP7, 5 and 3 genes and association studies in autism and schizophrenia in Japanese subjects. J Hum Genet 55, 127–130 (2010). https://doi.org/10.1038/jhg.2009.133
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.133
Keywords
This article is cited by
-
FABP7 Protects Astrocytes Against ROS Toxicity via Lipid Droplet Formation
Molecular Neurobiology (2019)
-
Investigation of the fatty acid transporter-encoding genes SLC27A3 and SLC27A4 in autism
Scientific Reports (2015)
-
Emerging roles of astrocytes in neural circuit development
Nature Reviews Neuroscience (2013)
-
The human fatty acid-binding protein family: Evolutionary divergences and functions
Human Genomics (2011)
-
Identification of FABP7 in fibroblastic reticular cells of mouse lymph nodes
Histochemistry and Cell Biology (2010)