Abstract
Deletion of exons 45–55 (del45–55) in the Duchenne muscular dystrophy gene (DMD) has gained particular interest in the field of molecular therapy, because it causes a milder phenotype than DMD, and therefore, may represent a good candidate for the goal of a multiple exon-skipping strategy. We have precisely characterized deletion breakpoints in three patients with del45–55 in DMD. Two of them were young adult males of the X-linked dilated cardiomyopathy phenotype, and the third patient revealed the mild Becker muscular dystrophy phenotype of late onset. The deletion breakpoints differed among patients. The deletion started at nt 226 604, 231 518, 117 284 in intron 44, and ended at nt 64 994, 59 314, 71 806 in intron 55, respectively. Deletion junctions showed no significant homology between the sequences adjacent to the distal and proximal end joints in these patients. Deletion breakpoints were not primarily associated with any particular sequence element, or with a matrix attachment region. However, there were several palindromic sequences and short tandem repeats at or near the breakpoints. These sequences, with a marked propensity to form secondary DNA structure intermediates, may predispose local DNA to breakage and intragenic recombination in these patients.
Similar content being viewed by others
Introduction
The Duchenne muscular dystrophy gene (DMD) is the largest one so far identified, spanning more than 2.5 Mb and occupying roughly 0.1% of the human genome.1 Mutations in DMD cause a devastating muscular dystrophy named Duchenne/Becker muscular dystrophy (DMD/BMD). The cardio-specific phenotype of dystrophinopathy is also known as X-linked dilated cardiomyopathy (XLDCM).1 Intragenic deletions and duplications together account for over two-thirds of the mutations found in DMD/BMD patients.2, 3, 4, 5, 6, 7 Despite heterogeneity in both deletion size and location, two deletion ‘hotspots’ have been identified in DMD: a minor ‘hotspot’ including exons 2–19, and the major one involving exons 40–50 or 45–55.2, 4, 5, 7
Among deletions of variable sizes and locations found in DMD/BMD patients, deletion of exons 45–55 (del45–55) in DMD has gained particular attention because an artificial dystrophin with del45–55 created by a multiple exon-skipping strategy could transform the DMD phenotype into the asymptomatic or milder BMD phenotype.8 We reported three patients with del45–55 who presented with the phenotypes far from DMD.9 Two of them were young adult males of the XLDCM phenotype.9, 10 The third patient revealed the mild BMD phenotype of late onset.9, 11 Our patients support the hypothesis that del45–55 could be a good candidate for the goal of a multiple exon-skipping strategy for DMD.
To elucidate the molecular basis of del45–55 in more detail, we have characterized the breakpoints of del45–55 in DMD in these three patients. To our knowledge, only two breakpoints have been precisely described in intron 44,12 which is the major deletion-prone region in DMD.1, 6, 7, 13 On the other hand, an analysis of breakpoints in intron 55 has never been reported.
Materials and methods
Patients
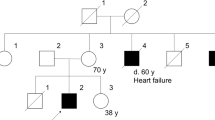
Clinical details of the patients were reported earlier.9, 10, 11 The patient number in this study corresponded to that in the previous report.9 Patients 1 and 2 shared the similar clinical phenotype of XLDCM. They had been free of muscular symptoms before they developed congestive heart failure along with cold-like symptoms at the age of 26 and 36 years, respectively.9, 10 Despite no obvious muscular atrophy or weakness, the hyperCKemia reminded us of dystrophinopathy. Patient 3 showed the mild BMD phenotype with muscular atrophy and weakness in the extremities, which began around the age of 59 years.9, 11 He showed no obvious cardiac involvement.
Identification of breakpoints
The sequences of introns 44 and 55 were derived from the University of California Santa Cruz (UCSC) human genome website (http://genome.ucsc.edu). The nucleotide numbering was started from the first nucleotide for each intron in this paper. A total of 53 primer sets were designed to roughly span introns 44 (28 sets) and 55 (25 sets), and to yield PCR products of 200–300 bp. Using these primer sets, three patients were independently screened by PCR for the presence or absence of regions targeted by primer sets. The failure of PCR amplification indicated that either the entire fragment or at least one annealing site was deleted. PCR reaction conditions were as follows: 95 °C for 1 min (initial denaturation), 30 cycles of 95 °C for 30 s (denaturation), 60 °C for 1 min (annealing), 72 °C for 1 min (extension) and 95 °C for 7 min (final extension). When mapping of both 5′ and 3′ breakpoints was restricted to less than 1–2 kb, direct amplification was tried to obtain the junctional fragments with a combination of sense primers in intron 44 and antisense primers in intron 55. As amplification of deletion junctions always yielded a single band, the PCR products were purified and sequenced.
Intron sequence analysis
Searches for interspersed repetitive elements were performed by using the RepeatMasker program (http://repeatmasker.genome.washington.edu, http://repeatmasker.org) run under sensitive settings (we selected ‘cross_match’ as search engine, and ‘slow’ as speed/sensitivity). Matrix attachment regions (MARs) were identified by using the MAR-Wiz program (http://www.futuresoft.org/modules/MarFinder/index.html). In MAR-Wiz analysis, all the rules included in the Core MAR rules were selected. Sliding window parameters were as follows: window width 1000, slide distance 100, cutoff threshold 0.6 and run-length 3.
Results
Sequence characterization of introns 44 and 55 in DMD
The sequences of introns 44 and 55 were first analyzed for interspersed repetitive elements, the frequency of which was 32. 7% for intron 44 and 40.5% for intron 55 (S1, Electric Supplementary material). The value for intron 55, but not for intron 44, was higher than the corresponding value for the whole DMD (35.6%).14
The distribution of repetitive elements and the possible MAR sites are schematically shown in (S2, Electric Supplementary material). There were four regions with a cutoff threshold of more than 0.6 in intron 44; they were the regions around nt 33 000 (average strength 0.658), nt 124 000 (0.715), nt 131 300 (0.749) and nt 174 800 (0.819). Those regions in intron 55 were as follows: the regions around nt 4700 (0.740) and nt 35 300 (0.758). The regions around nt 1100 in intron 44 and nt 58 200 in intron 55 were also found to have a relatively high MAR potential.
Analysis of breakpoint sequences
The sizes and breakpoints of the deletion were different in these patients (Figure 1), but the breakpoints of patients 1 and 2 were very close, relative to the large size of introns 44 and 55 (Figure 1). The deletions spanned 427.6 kb (patient 1), 417.1 kb (patient 2) and 515.5 kb (patient 3). None of the deletions affected the promoter regions and unique exon 1 of dystrophin isoforms Dp140 and Dp116, which are located in the introns 44 and 55, respectively.15, 16
Deletion breakpoints did not appear to be preferentially associated with any particular type of repetitive elements, although three of the six breakpoints were located in LINE-1 elements (S3, Electric Supplementary material). The breakpoints in intron 44 in these patients gathered in the 3′-half of that intron (Figure 1; S2, Electric Supplementary material), as pointed out earlier.6, 12, 13 Further the breakpoints in intron 55 showed a relative clustering within a region of approximately 10 kb (Figure 1; S2, Electric Supplementary material). None of the breakpoints appeared to have a close association with MAR sites.
Figure 2 shows the breakpoint sequences of the three patients compared with 40–50 bp of wild-type sequence on each side. Deletion junctions did not share a significant homology between the sequences adjacent to the distal and proximal end points in these patients. In patient 1, a 12-bp triplication was found at the breakpoint. The sequence TTTAAA, which is known to be capable of inducing a curvature of DNA, was present near the breakpoints of both introns 44 and 55 in patient 1 (Figure 2). Short (6–9 bp) tandem repeats were observed at or near the breakpoints in patients 1 and 2 (Figure 2). Palindromic sequences with 6 or more base pairs were found near the breakpoints in patient 1 (Figure 2). The 7-bp complementary sequences that flanked the breakpoint were found in intron 44 in patient 3.
Genomic sequences spanning the breakpoints in three patients, with the corresponding normal intronic regions. Only the 5′–3′ strands are shown. Triplicated regions are in white boxes, and homology regions across the deletion breakpoints are in gray boxes. Arrows and dotted arrows indicate palindromic sequences with a stem of 6 or more base pairs and short tandem repeats, respectively. Lines indicate runs of consecutive pyrimidine nucleotides. Dotted lines indicate the short deletion consensus sequence, TG(A/G)(A/G)(G/T)(A/C). Dots indicate the sequence TTTAAA.
Discussion
In this report, we have precisely characterized the breakpoints in three patients with del45–55 in DMD. We have detected no substantial homologies between the normal DNA sequences located across the breakpoints in introns 44 and 55. One of the breakpoints was located in LINE-1 repetitive elements in all patients, but the other ends were not associated with any particular repetitive elements. These observations in the patients with del45–55 are consistent with those in the cases with other types of deletion in DMD,12, 13, 14, 17, 18, 19 again supporting that homologous recombination is not the major cause of deletions in DMD.
Human gene deletions are induced by several mechanisms, the relative importance of which is probably governed by local and secondary DNA structures.20 Krawczak and Cooper20 have pointed out that human sequences involved in deletion events often contain a short deletion consensus sequence, TG(A/G)(A/G)(G/T)(A/C), and are often associated with palindromic sequences. Palindromic sequences are likely to locally induce the formation of a hairpin loop structure in a single strand of DNA, and serve as a strong topoisomerase II cleavage site.21, 22 We have identified several consensus sequences, palindromic sequences and short tandem repeats at or near the breakpoints. These sequences may promote DNA instability by facilitating the formation of secondary structure intermediates, which predispose DNA to breakage and intragenic recombination.17, 20, 23
The deletion breakpoints were different in three patients analyzed in this study; however, the breakpoints of patients 1 and 2 were in very close proximity in the relatively large introns 44 and 55. The difference of breakpoint locations between these two patients was approximately 5000 bp at both ends within introns 44 and 55. Their clinical features, compatible with XLDCM, were very similar to each other, and outstanding among dystrophinopathy. It is possible that the close similarity of clinical features and molecular defects has some pathogenic or biological significance. Some trans- or cis-acting elements that are involved in the sequence within or around the deleted regions may contribute to the pathogenesis of this unique phenotype in these patients. Accumulation of dystrophinopathy patients with del45–55 and further detailed analysis of the sequence in introns 44 and 55 will be needed to answer this hypothesis.
References
Muntoni, F., Torelli, S. & Ferlini, A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2, 731–740 (2003).
Forrest, S. M., Cross, G. S., Speer, A., Gardner-Medwin, D., Burn, J. & Davies, K. E. Preferential deletion of exons in Duchenne and Becker muscular dystrophies. Nature 329, 638–640 (1987).
Koenig, M., Hoffman, E. P., Bertelson, C. J., Monaco, A. P., Feener, C. & Kunkel, L. M. Cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50, 509–517 (1987).
Koenig, M., Beggs, A. H., Moyer, M., Scherpf, S., Heindrich, K., Bettecken, T. et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am. J. Hum. Genet. 45, 498–506 (1989).
den Dunnen, J. T., Grootscholten, P. M., Bakker, E., Blonden, L. A., Ginjaar, H. B., Wapenaar, M. C. et al. Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am. J. Hum. Genet. 45, 835–847 (1989).
Blonden, L. A., Grootscholten, P. M., den Dunnen, J. T., Bakker, E., Abbs, S., Bobrow, M. et al. 242 breakpoints in the 200-kb deletion-prone P20 region of the DMD gene are widely spread. Genomics 10, 631–639 (1991).
Oudet, C., Hanauer, A., Clemens, P., Caskey, T. & Mandel, J. L. Two hot spots of recombination in the DMD gene correlate with the deletion prone regions. Hum. Mol. Genet. 1, 599–603 (1992).
Béroud, C., Tuffery-Giraud, S., Matsuo, M., Hamroun, D., Humbertclaude, V., Monnier, N. et al. Multiexon skipping leading to an artificial DMD protein lacking amino acids from exons 45 through 55 could rescue up to 63% of patients with Duchenne muscular dystrophy. Hum. Mutat. 28, 196–202 (2007).
Nakamura, A., Yoshida, K., Fukushima, K., Ueda, H., Urasawa, N., Koyama, J. et al. Follow-up of three patients with a large deletion of exons 45–55 in the Duchenne muscular dystrophy (DMD) gene. J. Clin. Neurosci. 15, 757–763 (2008).
Tasaki, N., Yoshida, K., Haruta, S., Kouno, H., Ichinose, H., Fujimoto, Y. et al. X-linked dilated cardiomyopathy with a large hot-spot deletion in the dystrophin gene. Intern. Med. 40, 1215–1221 (2001).
Yazaki, M., Yoshida, K., Nakamura, A., Koyama, J., Nanba, T., Ohori, N. et al. Clinical characteristics of aged Becker muscular dystrophy patients with onset after 30 years. Eur. Neurol. 42, 145–149 (1999).
Love, D. R., England, S. B., Speer, A., Marsden, R. F., Bloomfield, J. F., Roche, A. L. et al. Sequences of junction fragments in the deletion-prone region of the dystrophin gene. Genomics 10, 57–67 (1991).
Sironi, M., Pozzoli, U., Cagliani, R., Giorda, R., Comi, G. P., Bardoni, A. et al. Relevance of sequence and structure elements for deletion events in the dystrophin gene major hot-spot. Hum. Genet. 112, 272–288 (2003).
Toffolatti, L., Cardazzo, B., Nobile, C., Danieli, G. A., Gualandi, F., Muntoni, F. et al. Investigating the mechanism of chromosomal deletion: characterization of 39 deletion breakpoints in introns 47 and 48 of the human dystrophin gene. Genomics 80, 523–530 (2002).
Byers, T. J., Lidov, H. G. W. & Kunkel, L. M. An alternative dystrophin transcript specific to peripheral nerve. Nat. Genet. 4, 77–81 (1993).
Lidov, H. G. W., Selig, S. & Kunkel, L. M. Dp140: a novel 140 kDa CNS transcript from the dystrophin locus. Hum. Mol. Genet. 4, 329–335 (1995).
McNaughton, J. C., Cockburn, D. J., Hughes, G., Jones, W. A., Laing, N. G., Ray, P. N. et al. Is gene deletion in eukaryotes sequence-dependent? A study of nine deletion junctions and nineteen other deletion breakpoints in intron 7 of the human dystrophin gene. Gene 222, 41–51 (1998).
Suminaga, R., Takeshima, Y., Yasuda, K., Shiga, N., Nakamura, H. & Matsuo, M. Non-homologous recombination between Alu and LINE-1 repeats caused a 430-kb deletion in the dystrophin gene: a novel source of genomic instability. J. Hum. Genet. 45, 331–336 (2000).
Nobile, C., Toffolatti, L., Rizzi, F., Simionati, B., Nigro, V., Cardazzo, B. et al. Analysis of 22 deletion breakpoints in dystrophin intron 49. Hum. Genet. 110, 418–421 (2002).
Krawczak, M. & Cooper, D. N. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum. Genet. 86, 425–441 (1991).
Froelich-Ammon, S. J., Gales, K. C. & Osheroff, N. Site-specific cleavage of a DNA hairpin by topoisomerase II. J. Biol. Chem. 269, 7719–7725 (1994).
Robinson, D. O., Bunyan, D. J., Gabb, H. A., Temple, I. K. & Yau, S. C. A small intraexonic deletion within the dystrophin gene suggests a possible mechanism of mutagenesis. Hum. Genet. 99, 658–662 (1997).
Spitzner, J. R., Chung, I. K. & Muller, M. T. Eukaryotic topoisomerase II preferentially cleaves alternating purine–pyrimidine repeats. Nucleic Acids Res. 18, 1–11 (1990).
Acknowledgements
This study was supported by a Research Grant for Nervous and Mental Disorders (8A-2) from the Ministry of Health and Welfare.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
Rights and permissions
About this article
Cite this article
Miyazaki, D., Yoshida, K., Fukushima, K. et al. Characterization of deletion breakpoints in patients with dystrophinopathy carrying a deletion of exons 45–55 of the Duchenne muscular dystrophy (DMD) gene. J Hum Genet 54, 127–130 (2009). https://doi.org/10.1038/jhg.2008.8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2008.8
Keywords
This article is cited by
-
Precise mapping of 17 deletion breakpoints within the central hotspot deletion region (introns 50 and 51) of the DMD gene
Journal of Human Genetics (2017)