Abstract
A considerable fraction of mutations associated with hereditary disorders and cancers affect splicing. Some of them cause exon skipping or the inclusion of an additional exon, whereas others lead to the inclusion of intronic sequences or deletion of exonic sequences through the activation of cryptic splice sites. We focused on the latter cases and have designed a series of vectors that express modified U7 small nuclear RNAs (snRNAs) containing a sequence antisense to the cryptic splice site. Three cases of such mutation were investigated in this study. In two of them, which occurred in the PTCH1 and BRCA1 genes, canonical splice donor sites had been partially impaired by mutations that activated nearby intronic cryptic splice donor sites. Another mutation found in exonic region in CYP11A created a novel splice donor site. Transient expression of the engineered U7 snRNAs in HeLa cells restored correct splicing in a sequence-specific and dose-dependent manner in the former two cases. In contrast, the third case, in which the cryptic splice donor site in the exonic sequence was activated, the expression of modified U7 snRNA resulted in exon skipping. The correction of aberrant splicing by suppressing intronic cryptic splice sites with modified U7 is expected be a promising alternative to gene replacement therapy.
Similar content being viewed by others
Introduction
Alternative splicing is one of the mechanisms that account for the greater macromolecular and cellular complexity of higher eukaryotic organisms (Ast 2004). In addition, splicing events are also implicated in pathophysiological processes, and it has been estimated that at least 15% of point mutations that cause human genetic diseases affect splicing (Krawczak et al. 1992). Specifically, in studies of neurofibromatosis type 1 and ataxia telangiectasia where analyses were performed at both the DNA and RNA levels, no less than 50% of the patients were found to have diseases due to mutations that resulted in aberrant splicing of the responsible genes, NF1 and ATM, respectively (Ars et al. 2003; Teraoka et al. 1999). Considering that multi-exon genes have a higher frequency of undergoing alternative splicing (up to 74%) (Johnson et al. 2003), larger genes are thought to be more prone to harboring mutations that affect splicing. In terms of gene therapy, these genes are generally difficult to replace by a large transgene because of the limitations of vectors suitable for efficient delivery. In addition to gene replacement therapy, gene therapy may also be accomplished by the manipulation of a gene’s structure and expression. Attempts have been made to alter mRNA structures in order to correct a limited number of genetic disorders such as Duchenne muscular dystrophy, spinal muscular atrophy and β-thalassemia (Goyenvalle et al. 2004; Madocsai et al. 2005; Gorman et al. 1998). In these studies, antisense sequences linked to a modified U7 small nuclear RNA (snRNA) have been constructed and delivered. U7, a snRNA molecule normally involved in processing of the histone mRNA 3′ end, can be engineered to bind appropriate Sm proteins, redistributed to the spliceosome, and used to deliver antisense sequences (Grimm et al. 1993). In these attempts, antisense sequences were designed to skip the mutation-containing exon (Goyenvalle et al. 2004), the additional exon generated by the splice site created by the mutation (Gorman et al. 1998) or internal exon of HIV-1 regulatory genes (Asparuhova et al. 2007). In another study, the production of a biologically active protein isoform was successfully increased by suppressing a 3′ splice site with an engineered U7 snRNA (Madocsai et al. 2005). Recently, we and others noticed that a considerable fraction of mutations disrupt authentic splice sites leading to the activation of cryptic splice sites (Roca et al. 2003; Nagao et al. 2005a). We also noticed that, in some of these cases, the authentic splice sites are only partially impaired and still have reasonably high consensus values for the splice sites. Therefore, we reasoned that, in these cases, inhibition of the cryptic sites using engineered U7 snRNA could possibly restore the normal splicing pattern. In this paper, we show that some of the aberrant splicings using intronic 5′ cryptic splice sites activated by the incomplete impairment of the canonical 5′ splice sites can be successfully corrected by engineered U7 snRNA.
Materials and methods
Constructs
The plasmid, pU7SmOPT, was kindly provided by Dr. D. Schümperli (Grimm et al. 1993). PCR-mediated mutagenesis was performed as described previously (Imai et al. 1991) using pU7SmOPT as a template. The primers used are listed in the supplementary Table. To generate minigene constructs for PTCH1 and BRCA1, genomic DNA was amplified by PCR with the primers listed in the supplementary Table. Recognition sequences for restriction enzymes were added to the 5′ ends of primers to facilitate subcloning. After the digestion of PCR products with appropriate restriction enzymes, PCR products were subcloned into pcDNA3.0. Minigenes for CYP11A were described previously (Katsumata et al. 2002). The authenticity of all constructs was confirmed by sequencing.
Cell culture and transfections
HeLa cells were grown in DMEM medium supplemented with 10% heat-inactivated fetal calf serum, 50 U/ml penicillin, and 0.1 mg/ml streptomycin. HeLa cells growing on 6-cm plates were transfected with 1 μg of plasmid DNA as indicated in the figure legends using Effectene reagent (Qiagen).
RT-PCR
Forty-eight hours after the transfection, total RNA was extracted using the RNeasy kit from Qiagen according to the manufacturer’s recommendations. RT-PCR was performed as previously described using 5 μg of total RNA (Nagao et al. 2005b). Primers used for RT-PCR are listed in the supplementary Table. To quantify the splicing, 1 μl of logarithmically amplifying PCR product was applied onto a DNA LabChip (Agilent Technologies) and loaded into an Agilent 2100 bioanalyzer. Data analysis was performed with Agilent 2100 bioanalyzer software, and the percentage of each splicing product based on molarity was calculated. Gel electrophoretograms were taken after ethidium bromide staining of 2% agarose gels or silver staining (Bio-Rad) of 3.5% polyacrylamide gels.
Results
Disruption of cryptic splice sites restores canonical splice sites in minigenes
PTCH1 is a gene responsible for nevoid basal cell carcinoma syndrome (NBCCS) characterized by minor developmental anomalies and an increased incidence of cancers such as medulloblastoma and basal cell carcinoma (Gorlin 1987; Hahn et al. 1996; Johnson et al. 1996). We previously reported a mutation, c.584G > A, in the PTCH1 gene (as per Genbank entry NM_000264.2: the A of the ATG of the initiator Met codon is counted as nucleotide +1) (Nagao et al. 2005a). This mutation found in patient G17 is located at the 3′ end of exon 3 and leads to an aberrant splicing in which a cryptic splice donor site located in intron 3 was activated. The resultant mRNA with an insertion of a 37-bp intronic sequence between exon 3 and exon 4 leads to premature termination of the PTCH1 protein (Fig. 1a). However, we suspected that the mutation would only partially impair the splice donor site because the deoxyguanosine at the 3′ end of exons is not necessarily conserved in human genes. Indeed, when the sequence around the 5′ splice site was investigated with a prediction program based on the algorithm by Shapiro et al. (Shapiro and Senapathy 1987), the mutated sequence still had a high score for a splice donor (Fig. 1b). Therefore, we next addressed the question of whether the inactivation of the cryptic splice donor in intron 3 can restore the authentic splicing pattern. For this purpose, a genomic sequence containing exon 3, intron 3, and exon 4 was subcloned into pcDNA3.0. The sequence of the same region harboring the G17 mutation with or without an additional artificial mutation that inactivates the cryptic splice site was also included in this study (Fig. 2a, G17 and G17 + mt). HeLa cells were transfected with these minigene constructs and transcripts from the constructs were amplified by RT-PCR. The forward primer was set on the plasmid sequence so that the endogenous PTCH1 transcript would not be amplified (Fig. 2a, top). As demonstrated previously in G17 genomic DNA (Nagao et al. 2005a), a larger PCR product was generated from the plasmid construct carrying the G17 mutation (Fig. 2b, lane 2). DNA sequencing confirmed the presence of an additional 37-bp intronic sequence, implying that this experiment using minigene constructs reflects splicing events in endogenous PTCH1. As anticipated, authentic splicing was restored by mutating the cryptic splice donor site even in the presence of the G17 mutation (Fig. 2b, lane 3).
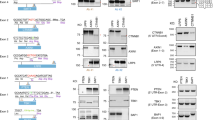
Aberrant splicing observed in an NBCCS patient, G17. a Schematic representation of the abnormal splicing identified in G17. The position of the point mutation is indicated by an asterisk. b Splice donor score profiles. Donor score profiles were obtained from the website supported by Deprtment of Genetics, The Hospital for Sick Children (http://www.violin.genet.sickkids.on.ca/∼ali/splicesitescore.html). The sequence encompassing exon 3 and intron 3 was scanned for the splice donor score with an 8-bp window. Ordinate represents calculated donor score. The mutated allele of G17 with or without an additional mutation at the cryptic donor site (see also Fig. 2a) was also subjected to this analysis. The arrow in the top panel indicates the authentic splice donor site while the one in the middle panel shows the cryptic splice donor site activated by the G17 mutation
Disruption of the activated cryptic splice donor site restores authentic splicing. a Minigene sequences used in the experiments. A schematic representation of the minigene is shown at the top. Arrows indicate the positions of the primers used for RT-PCR. The wild-type minigene (WT) carries exon 3, intron 3 and exon 4 of PTCH1. The mutated sequence was obtained from G17 genomic DNA. An additional mutation, GT→CA, was introduced in G17 to destroy the cryptic donor site (G17 + mt). b Correction of aberrant splicing observed in G17. HeLa cells were transfected with 1 μg of the minigene construct indicated at the top. Total RNA was extracted 48 h after the transfection and RT-PCR was performed using a pair of primers designed based on the vector sequence and on exon 4. The identities of spliced forms are indicated on the right
U7 snRNAs containing an anti-PTCH1 sequence modulate PTCH1 pre-mRNA splicing
The result described above prompted us to attempt inactivation of the cryptic splice site with U7 snRNA-based vectors. U7 snRNA forms a small nuclear particle consisting of an RNA component and several proteins. The 62-bp-long U7 RNA contains a region complementary to the histone pre-mRNA to initiate histone pre-mRNA 3′ end processing. By changing the wild-type Sm-binding site of U7 RNA into the consensus Sm-binding site (SmOpt), expression of U7 RNA is increased, while the activity for 3′ processing of the histone pre-mRNA is decreased (Grimm et al. 1993). Because U7 SmOpt-derived snRNA particles do not recruit the histone 3′-end-processing machinery, they can be used to target other pre-mRNAs by changing a portion complementary to the histone pre-mRNA. This approach has been proven successful in modulating pre-mRNA splicing in a number of applications (Goyenvalle et al. 2004; Madocsai et al. 2005; Gorman et al. 1998). Using a similar approach we have exchanged the natural anti-histone portion of U7 SmOpt RNA with a sequence complementary to the cryptic splice site of PTCH1 intron 3. Three modified anti-PTCH1 U7 snRNAs were generated to evaluate whether embedding antisense sequences against the PTCH1 pre-mRNA within U7 snRNA modulates aberrant splicing in G17 PTCH1 (Fig. 3a). When HeLa cells were transfected with these U7 snRNAs together with the minigene carrying the G17 mutation, a smaller PCR product that was identical in size to the authentic splicing product was generated indicating the correction of aberrant splicing (Fig. 3b, lanes 1–3). The proper splicing was confirmed by DNA sequencing. Transfection of U7 SmOpt, the plasmid carrying the natural anti-histone sequence at the 5′ end of the U7 snRNA, did not lead to any appreciable changes in the splicing pattern in G17 (Fig. 3b, lane 4), implying the specific interaction of the modified U7 snRNA with the PTCH1 target site. However, the effectiveness of the correction depended on the complementary sequence embedded in the plasmid and G17-2 carrying a cryptic splicing site at the center of the sequence was most effective in correcting aberrant splicing. Subsequently, titration experiments were performed by transfecting the most effective snRNA identified in the previous experiment (G17-2). Aberrant splicing was corrected dose dependently, and maximal modulation efficiency (51%) was achieved with the transfection of 0.95 μg of the U7 vector (Fig. 3c, lane 1). No further correction was obtained with higher amounts of the U7 vector transfected (data not shown).
Engineered U7 snRNAs modulate pre-mRNA splicing. a Schematic diagram of modified U7 snRNAs. Gray and black boxes represent the anti-PTCH1 and Sm protein binding sequences, respectively. The natural histone interacting sequence was exchanged for various anti-PTCH1 sequences targeting the cryptic splice donor site. b Representative agarose gel electrophoretogram of RT-PCR analysis and quantification. HeLa cells were transfected with 100 ng of the minigene construct together with 900 ng of one of the U7 constructs shown at the top. Total RNA was extracted 48 h after the transfection, and RT-PCR was performed using a pair of primers designed based on the vector sequence and on exon 4. The identities of the spliced forms are indicated on the right. The percentages of the authentic (corrected) splice form are shown at the bottom. c Dose dependence of anti-PTCH1 U7 snRNA transfection. HeLa cells were transfected with various combinations of U7 snRNA plasmids and minigenes as indicated at the top. The amounts of transfected DNAs are indicated in micrograms. The total amount of transfected DNA was adjusted to 1 μg. The percentages of the authentic splice form are shown at the bottom
U7 snRNAs containing an anti-BRCA1 sequence also modulate BRCA1 pre-mRNA splicing
In order to demonstrate that our strategy is applicable to other mutations, a similar mutation in the BRCA1 gene was employed for the next experiment. The mutation, IVS16 + 6T > C, found in a patient with breast cancer, was located in intron 16 of BRCA1 (Scholl et al. 1999), the tumor suppressor breast/ovarian cancer susceptibility gene (Miki et al. 1994). This mutation leads to a 65-bp insertion from the 5′ end of intron 16 due to the activation of the cryptic splice donor site (Fig. 4a), resulting in a premature termination of the BRCA1 protein. Plasmids that express engineered U7 snRNAs were then produced as described earlier by exchanging the anti-histone portion with one of the three complementary sequences encompassing the activated cryptic splice site in intron 16 (Fig. 4b). In order to know the best construct of the three, HeLa cells were transfected with each of the three U7 plasmids in combination with the minigene construct and the effect on splicing was analyzed. Although less efficiently than with the G17 plasmid, aberrant splicing was partially, but significantly corrected by one of the engineered snRNA constructs, BRCA1-1 (Fig. 4c, lane 1). Again, aberrant splicing was corrected in a dose-dependent fashion when various amounts of BRCA1-1 were used for transfection and maximal modulation efficiency was ~10% (Fig. 4d).
Correction of BRCA1 pre-mRNA splicing. a Schematic representation of abnormal splicing identified in IVS16 + 6T > C. The position of the point mutation is indicated by an asterisk. b Schematic diagram of modified U7 snRNAs. The natural histone interacting sequence was exchanged for various anti-BRCA1 sequences targeting the cryptic splice donor site. c Representative polyacrylamide gel electrophoretogram of RT-PCR analysis and quantification. Transfection and RT-PCR were performed as described in Fig. 3b. The identities of the spliced forms are indicated on the right. The percentages of the authentic (corrected) splice form are shown at the bottom. d Dose dependence of anti-BRCA1 U7 snRNA transfection. HeLa cells were transfected with various combinations of U7 snRNA plasmids and minigenes as indicated at the top. The amounts of transfected DNAs are indicated in micrograms. The total amount of transfected DNA was adjusted to 1 μg. The percentages of the authentic splice form are shown at the bottom
Cryptic splice donor sites that lie in the exonic sequence are intractable to U7-mediated correction
We next focused on another type of splicing mutation, which creates a cryptic splice site (de novo splice donor site). The mutation, c.566C > T, found in a patient with congenital adrenal insufficiency, was located in exon 3 of CYP11A (Katsumata et al. 2002), which encodes a cholesterol side-chain cleavage enzyme (Morohashi et al. 1987). This mutation has been demonstrated to create a consensus splice donor sequence and causes an aberrant splicing resulting in a 61-bp deletion rather than a missense mutation (Katsumata et al. 2002) (Fig. 5a). As described above, modified U7 snRNA plasmids were constructed in which various antisense sequences targeted at the activated cryptic donor site (Fig. 5b). When HeLa cells were transfected with these plasmids together with the minigene construct, the splicing pattern was significantly altered. However, unexpectedly, the targeted exon was deleted to various degrees (Fig. 5c, lanes 1–3, c), rather than the authentic splicing being restored (Fig. 5c, lanes 1–3, a). This may due to suppression of an exonic splicing enhancer (ESE), because potential motifs for SR proteins, SC35 and SF2/ASF, were found adjacent to the targeted sequence using ESEfinder (http://www.rulai.cshl.edu/tools/ESE/) (Cartegni et al. 2003) (Fig. 5b, solid and broken lines, respectively).
Modification of CYP11A splicing with anti-CYP11A U7 snRNA. a Schematic representation of abnormal splicing identified in 566C > T (as per GenBank entry NM_000781: the A of the first ATG of the initiator Met codon is counted as nucleotide +1). The position of the point mutation is indicated by an asterisk. b Schematic diagram of modified U7 snRNAs. The natural histone-interacting sequence was exchanged for various anti-CYP11A sequences targeting the cryptic splice donor site. Potential motifs for SC35 and SF2/ASF are indicated by solid and broken lines, respectively (see text). c Representative agarose electrophoretogram of RT-PCR analysis and quantification. Transfection and RT-PCR were performed as described in Fig. 3b. The identities of the spliced forms are indicated on the right. The percentages of each spliced product are shown at the bottom
Discussion
Despite the tremendous progress in gene replacement therapy, its clinical use is still limited mainly due to low efficiency of gene transfer, inability to deliver large genes, and lack of endogenous regulatory control. To circumvent these obstacles, the modified U7 gene, along with its natural promoter and 3′ elements, has been used to modify the mRNA maturation process as a potential alternative to gene replacement therapy. Thus far, attempts have been made to modulate inclusion/exclusion types of splice alterations (Goyenvalle et al. 2004; Madocsai et al. 2005; Gorman et al. 1998; Asparuhova et al. 2007). Although the most common outcome of mutations affecting splice sites is exon skipping, the activation of cryptic splice sites is ranked second (Krawczak et al. 1992; Nakai and Sakamoto 1994). Our study is unique in that modified snRNAs are targeted at the cryptic splice site activated by incomplete disruption of authentic splice site. A considerable number of splice mutations take place outside of the conserved AG acceptor dinucleotide or GT donor nucleotide (Krawczak et al. 1992; Kralovicova et al. 2005) implying that the disruption of the splice sequence is incomplete in these cases. In addition, such mutations tend to be overlooked unless they are investigated at RNA level. Therefore, our strategy is potentially widely applicable. From our experiment, intronic cryptic donor sites are relatively easy to suppress by U7 snRNAs. In contrast, although more cases need to be investigated, the suppression of exonic cryptic sites may result in skipping of the exon in which they are located depending on the presence of the nearby ESE as in CYP11A 566C > T. Exon skipping in a similar situation has been reported by Vacek et al. (2003). They have used a U7 snRNA targeting an exon-internal location to induce exon skipping in a thalassemic HBB gene.
The effects of engineered U7 snRNA are expected to be limited only to cells that express the target sequence for the following reasons. First, the GenBank database of human sequences contains no sequences other than respective genes that correspond to the antisense sequence used for our study, even allowing for two mismatches. Of note, oligonucleotide with a two-nucleotide mismatch gave no antisense effect (Sierakowska et al. 1996). Second, neither of the plasmids caused any detectable changes in the morphology or growth rate of transfected cells (Sierakowska et al. 1996; Vacek et al. 2003; data not shown). However, it may be intriguing to see the influence of engineered U7 snRNA on cellular splicing using exon junction microarrays (Johnson et al. 2003; Nagao et al. 2005a).
Although the U7 construct has to be constructed on a case-by-case basis, this technology can potentially be applied to a wide variety of diseases regardless of the size of the responsible genes. This strategy is particularly useful for large genes because a full-length cDNA is difficult to introduce in these cases. Another benefit of this type of approach is that it can be used for the disorders that develop from gain-of-function mutations including dominant negative ones as well as loss-of-function mutations. In the former type of diseases, introduction of the wild-type cDNA is unlikely to alleviate phenotypes.
In summary, we have demonstrated that aberrant splicing using cryptic splice donor sites activated by incomplete impairment of the authentic donor site can be successfully corrected with U7-based vectors by suppressing the cryptic sites. Although the effectiveness of U7 snRNA should be evaluated in each mutation, viral gene delivery system integrating modified U7 snRNAs is expected to become a promising form of gene therapy in the future.
References
Ars E, Kruyer H, Morell M, Pros E, Serra E, Ravella A, Estivill X, Lazaro C (2003) Recurrent mutations in the NF1 gene are common among neurofibromatosis type 1 patients. J Med Genet 40:e82
Asparuhova MB, Marti G, Liu S, Serhan F, Trono D, Schümperli D (2007) Inhibition of HIV-1 multiplication by a modified U7 snRNA inducing Tat and Rev exon skipping. J Gene Med 9:323–334
Ast G (2004) How did alternative splicing evolve? Nat Rev Genet 5:773–782
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31:3568–3571
Gorlin RJ (1987) Nevoid basal-cell carcinoma syndrome. Medicine (Baltimore) 66:98–113
Gorman L, Suter D, Emerick V, Schümperli D, Kole R (1998) Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc Natl Acad Sci USA 95:4929–4934
Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L, Danos O (2004) Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science 306:1796–1799
Grimm C, Stefanovic B, Schümperli D (1993) The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J 12:1229–1238
Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, Pressman C, Leffell DJ, Gerrard B, Goldstein AM, Dean M, Toftgard R, Chenevix-Trench G, Wainwright B, Bale AE (1996) Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85:841–851
Imai Y, Matsushima Y, Sugimura T, Terada M (1991) A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res 19:2785
Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141–2144
Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH Jr, Scott MP (1996) Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272:1668–1671
Katsumata N, Ohtake M, Hojo T, Ogawa E, Hara T, Sato N, Tanaka T (2002) Compound heterozygous mutations in the cholesterol side-chain cleavage enzyme gene (CYP11A) cause congenital adrenal insufficiency in humans. J Clin Endocrinol Metab 87:3808–3813
Kralovicova J, Christensen MB, Vorechovsky I (2005) Biased exon/intron distribution of cryptic and de novo 3′ splice sites. Nucleic Acids Res 33:4882–4898
Krawczak M, Reiss J, Cooper DN (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet 90:41–54
Madocsai C, Lim SR, Geib T, Lam BJ, Hertel KJ (2005) Correction of SMN2 Pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol Ther 12:1013–1022
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66–71
Morohashi K, Sogawa K, Omura T, Fujii-Kuriyama Y (1987) Gene structure of human cytochrome P-450(SCC), cholesterol desmolase. J Biochem (Tokyo) 101:879–887
Nagao K, Togawa N, Fujii K, Uchikawa H, Kohno Y, Yamada M, Miyashita T (2005a) Detecting tissue-specific alternative splicing and disease-associated aberrant splicing of the PTCH gene with exon junction microarrays. Hum Mol Genet 14:3379–3388
Nagao K, Toyoda M, Takeuchi-Inoue K, Fujii K, Yamada M, Miyashita T (2005b) Identification and characterization of multiple isoforms of a murine and human tumor suppressor, patched, having distinct first exons. Genomics 85:462–471
Nakai K, Sakamoto H (1994) Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene 141:171–177
Roca X, Sachidanandam R, Krainer AR (2003) Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res 31:6321–6333
Scholl T, Pyne MT, Russo D, Ward BE (1999) BRCA1 IVS16 + 6T–>C is a deleterious mutation that creates an aberrant transcript by activating a cryptic splice donor site. Am J Med Genet 85:113–116
Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174
Sierakowska H, Sambade MJ, Agrawal S, Kole R (1996) Repair of thalassemic human β-globin mRNA in mammalian cells by antisense oligonucleotides. Proc Natl Acad Sci USA 93:12840–12844
Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, Chessa L, Sanal O, Bernatowska E, Gatti RA, Concannon P (1999) Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet 64:1617–1631
Vacek MM, Ma H, Gemignani F, Lacerra G, Kafri T, Kole R (2003) High-level expression of hemoglobin A in human thalassemic erythroid progenitor cells following lentiviral vector delivery of an antisense snRNA. Blood 101:104–111
Acknowledgments
We are grateful to Dr. Daniel Schümperli for providing U7 SmOPT. We thank Mayu Yamazaki-Inoue for her technical support. This work was supported by the Naito Foundation and Grants for Cancer Research from the Ministry of Health, Labour and Welfare and a Grant-in-Aid for Scientific Research and the Budget for Nuclear Research from the Ministry of Education, Culture, Sports, Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10038_2007_192_MOESM1_ESM.doc
Supplementary Table List of primers used in this study. Restriction sites added to facilitate subcloning are underlined. (DOC 50 kb)
Rights and permissions
About this article
Cite this article
Uchikawa, H., Fujii, K., Kohno, Y. et al. U7 snRNA-mediated correction of aberrant splicing caused by activation of cryptic splice sites. J Hum Genet 52, 891–897 (2007). https://doi.org/10.1007/s10038-007-0192-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-007-0192-8