Abstract
Airway inflammation is a major factor in the pathogenesis of asthma. Interleukin 8 (IL8) is a potent proinflammatory cytokine that interacts with its receptors, IL8RA and IL8RB. We investigated the genetic polymorphisms in IL8, IL8RA, and IL8RB for any association with risk of asthma and peripheral blood eosinophil counts in a Korean population. By carrying out direct sequencing in 24 individuals, we identified 20 sequence variants within exons and their flanking regions, including the 1.5 kb promoter regions of IL8, IL8RA, and IL8RB. Among them, seven common single-nucleotide polymorphisms (SNPs) were selected for genotyping in our asthma cohort (n = 1,439). Two common haplotypes in IL8 and three in IL8RA and IL8RB (defined as one block) were identified. Although none of the polymorphisms showed a significant association with risk of asthma, IL8RA-B ht2 showed a significant association with the peripheral blood eosinophil counts (%) among asthma patients, e.g., lower eosinophil levels among individuals with the homozygous IL8RA-B ht2 (3.55 ± 3.39%) than among other asthmatic patients (5.52 ± 5.55%; P corr = 0.018). Our findings suggest that polymorphisms and haplotypes in IL8RA and IL8RB might be among the genetic factors underlying production of peripheral blood eosinophil.
Similar content being viewed by others
Introduction
Cytokines and their receptors play an important role in the coordination and persistence of the inflammatory process in the chronic inflammation of the airways in asthma and many other diseases. Chronic and acute inflammatory changes observed in the asthmatic airway may result from the excessive release of many types of cytokines (Bentley et al. 1992; Broide et al. 1992; Chung and Barnes 1999). Eosinophils have the potential to synthesize and release a number of cytokines and chemokines. Cytokines produced by eosinophils include the autocrine-eosinophil active growth factor (IL3, IL5, and GM-CSF), immunoregulatory cytokines (IL2, IL4, IL1, TGFB, and IFNG), and proinflammatory cytokines (IL1, IL6, TNFA, and IL16) and chemokines (IL8, MIP-1α, and RANTES; Desreumaux and Capron 1996).
Among the proinflammatory cytokines and chemokines, IL8, on chromosome 4q13–q21, is mainly involved in the initiation and amplification of acute inflammatory reaction and in the chronic inflammatory process (Harada et al. 1994). The important role of IL8 in the pathophysiology of bronchial asthma has been confirmed by many studies in humans. Increased levels of IL8 in sputum precede an exacerbation of asthma, and IL8-producing cells are more frequently found in non-atopic asthma patients. In addition, IL8 selectively inhibits IgE production in atopic patients (Amin et al. 2000; Heinzmann et al. 2004; Kurashima et al. 1996). Recently, several studies have reported the association of the polymorphism −251T within the promoter of IL8 with respiratory syncytial virus (RSV) bronchiolitis in the UK, and juvenile idiopathic arthritis and asthma in Germany (Heinzmann et al. 2004, Hull et al. 2000, 2001).
IL8 mediates chemoattraction via two different receptors: IL8RA and IL8RB. IL8RA and IL8RB are members of the G-protein-coupled receptor family and these two receptors have 77% amino acid identity. IL8RA and IL8RB bind to IL8 with high affinity, and transduce the signal through a G-protein-activated second messenger system. IL8RA is more specific for IL8 and will bind other cytokines with much lower affinity, while IL8RB is less specific for IL8 (Lee et al. 1992). IL8RA and IL8RB form a gene cluster in a region mapped to chromosome 2q35. In a recent study, one non-synonymous SNP in IL8RA showed significant association with chronic obstructive pulmonary disease (COPD) and asthma in a German cohort (Stemmler et al. 2005). Moreover, the chromosome region in which IL8RA and IL8RB lie revealed the highest linkage to spirometric phenotypes for early-onset COPD in genome-wide screening (Palmer et al. 2003). The same region also showed evidence of linkage to total serum IgE levels in asthma patients (Xu et al. 2000).
Based on the biological properties and previous association studies of asthma and related phenotypes as above, it is hypothesized that IL8, IL8RA, and IL8RB play an important role in the development of asthma in the Korean population. We performed extensive screening of IL8, IL8RA, and IL8RB by carrying out direct sequencing to detect additional polymorphisms and examined the genetic association with the risk of asthma and eosinophil counts. Here, we present 20 genetic polymorphisms found in IL8, IL8RA, and IL8RB and the results of an association study in a Korean asthma cohort.
Materials and methods
Participants
Participants were recruited from the Asthma Genome Research Center, which consists of four tertiary hospitals in Korea (Soonchunhyang University Hospital, Ajuo University Hospital, Ulsan University Hospital, and Choong-Ang University Hospital). Ethical approval was obtained from the institutional review board of each hospital. All patients had the clinical symptoms and the physical examination results compatible with asthma. Each patient showed airway reversibility as documented by an inhalant bronchodilator-induced improvement of more than 15% of forced expiratory volume in 1 s (FEV1) and/or an airway hyperreactivity of less than 10 mg/ml of methacholine. Normal participants were recruited from spouses of the patients and the general population who answered negatively to a screening questionnaire for respiratory symptoms and had FEV1 greater than the 75% predicted, the provocation concentration causing a fall in the FEV1 of 20% (PC20) by more than 10 mg/ml of methacholine, and normal findings on a simple chest radiogram. Total IgE and specific IgE to Dermatophagoides farinae (D.f.) and D. pteronyssinus (D.p.) were measured using the UniCAP system (Pharmacia Diagnostics, Sweden). Atopy was defined as having wheal reaction to allergen extract equal to or greater than that to histamine (1 mg/ml) or 3 mm in diameter. The peripheral blood eosinophil counts were measured using a Coulter GenS Hematology Analyzer (Miami, FL, USA). The clinical parameters are summarized in Table 1.
Sequencing analysis of IL8, IL8RA, and IL8RB
We sequenced all exons of the three genes (IL8, IL8RA, and IL8RB) and their flanking regions, including promoter regions (1.5 kb), to discover sequence variants in 24 unregulated individual Korean DNA samples using the ABI PRISM 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). Twenty-two primer sets for the amplification and sequencing analysis were designed based on GenBank sequences (Ref. genome sequences IL8: NT_006216, IL8RA, and IL8RB: NT_005403). Information regarding primers is available on our website (http://www.snp-genetics.com/reference). Sequence variants were verified by chromatograms.
Genotyping with fluorescence polarization detection
For genotyping of polymorphic sites, amplifying primers and probes were designed for TaqMan (Livak 1999). Primer Express (Applied Biosystems) was used to design both the PCR primers and the MGB TaqMan probes. One allelic probe was labeled with the 6-carboxy-fluorescein (FAM) dye and the other with the fluorescent VIC dye. Typically, PCR was run in the TaqMan Universal Master mix without UNG (Applied Biosystems) at a primer concentration of 900 nM and TaqMan MGB probe concentration of 200 nM. The reaction was performed in a 384-well format in a total reaction volume of 5 μl using 20 ng of genomic DNA. The plate was then placed in a thermal cycler (PE 9700; Applied Biosystems) and heated for 2 min at 50°C and for 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The TaqMan assay plate was then transferred to a Prism 7900HT instrument (Applied Biosystems) where the fluorescence intensity of each well was read. Fluorescence data files from each plate were analyzed by automated software (SDS 2.1). Detailed information concerning the primers can be obtained at the website mentioned above. For comparison of frequencies, haplotypes, and linkage disequilibrium coefficients with other major ethnic groups, we also genotyped 50 Caucasian and 50 African-American DNAs obtained from the Human Genetic Cell Repository (http://www.locus.umdnj.edu/nigms/).
Statistics
The χ2 tests were used to determine whether individual variants were in equilibrium at each locus in the population (Hardy–Weinberg equilibrium). We employed a widely used measure of linkage disequilibrium between all pairs of biallelic loci, Lewontin’s D′ (|D′|; Hedrick 1987) and r 2. Haplotypes of each individual were inferred using software (PHASE) based on the algorithm developed by Stephens et al. (2001), which uses a Bayesian approach that incorporates a priori expectations of haplotypic structures from population genetics and coalescent theory. Genotype distributions of IL8, IL8RA, and IL8RB polymorphisms and haplotypes between asthmatics and normal participants, and between atopic and non-atopic participants were analyzed with logistic regression models controlling for age (continuous value), sex (male = 0, female = 1), and smoking status (non-smoker = 0, ex-smoker = 1, smoker = 2) as covariates. Means and standard deviations (SD) of total serum IgE level (log transformed) and peripheral blood eosinophil counts (%) and P values were calculated using multiple regression analyses controlling for age, sex, and smoking status as covariates as above. P values of haplotype associations were calculated by the algorithm developed by Schaid et al. (2002; Haplo.Score). The effective number of independent marker loci in IL8, IL8RA, and IL8RB was calculated to correct for multiple testing. The effective number was calculated using the software SNPSpD (http://www.genepi.qimr.edu.au/general/daleN/SNPSpD/), which is based on the spectral decomposition (SpD) of matrices of pair-wise linkage disequilibrium (LD) between SNPs. The resulting number of independent marker loci was applied to correct for multiple testing (Nyholt 2004).
Results
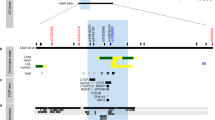
By direct sequencing in 24 individuals, we identified 20 sequence variants within exons and their flanking regions, including the 1.5 kb promoter regions of IL8, IL8RA, and IL8RB (Ad. Fig. 1). Among them, seven common SNPs [IL8 −352T > A, IL8 +2670A > T, IL8RA +92T > G (Met31Arg), IL8RA +827G > C (Ser276Thr), IL8RB −8520G > A, IL8RB +768C > T (Val256Val), and IL8RB +786C > T (Leu262Leu)] were selected for genotyping based on locations (SNPs in exons were preferred), LDs (only one SNP if there were absolute LDs (r2 = 1)), and frequencies (> 0.05) in our asthma cohort. The minor allele frequencies of genotyped polymorphisms were 0.348 (IL8 −352T > A), 0.333 (IL8 +2670A > T), 0.011 (IL8RA+92T > G), 0.087 (IL8RA +827G > C), 0.373 (IL8RB −8520G > A), 0.020 (IL8RB +768C > T), and 0.350 (IL8RB +786C > T) in the Korean population (n = 1,439; Ad. Fig. 1, Ad. Table 1). Genotype distributions of all polymorphisms in this study were in Hardy–Weinberg equilibrium (P > 0.05; Ad. Table 1).
Pair-wise comparison between two SNPs (−352T > A and +2670A > T) in IL8 revealed almost absolute LD (|D′| = 1, r2 = 1; Ad. Fig. 1A3), and two major haplotypes in IL8 were constructed in the Korean population using PHASE software (Stephens et al. 2001; Ad. Fig. 1A2). Haplotype analyses were not performed because ht1 and ht2 of IL8 were almost completely tagged by −352T > A and +2670A > T respectively. Haplotype 3 of IL8 was not analyzed either due to low frequency in Koreans. IL8RA and IL8RB form a gene cluster on chromosome 2q35 (25.5 kb apart), therefore, pair-wise comparison and haplotype construction were performed as a single LD block. As a result, several complete LDs (|D′| = 1 and r2 ≠ 1) were detected and three common haplotypes (frequency > 0.05) were constructed with five SNPs (IL8RA +92T > G, IL8RA +827G > C, IL8RB −8520G > A, IL8RB +768C > T, and IL8RB +786C > T) in Korean (Ad. Fig. 1B2 and B3). IL8RA-Bht1 and IL8RA-Bht3 were excluded from further analyses because they were almost completely tagged by IL8RB −8520G > A and IL8RA +827G > C respectively. Haplotype frequencies and LDs were compared among three ethnic groups (Ad. Fig. 1).
Allele frequencies of each SNP and IL8RA-Bht2 were compared among asthma patients and normal controls using a logistic regression model controlling for age, sex, and smoking status as covariates. None of the polymorphisms in IL8, IL8RA, and IL8RB showed a significant association with risk of asthma (Table 2). Also, no significant associations were detected with atopic status and total serum IgE level (data not shown).
In further analysis, associations of IL8, IL8RA, and IL8RB polymorphisms with peripheral blood eosinophil counts (%) were analyzed using regression models controlling for age, sex, and smoking status as covariates. The obtained P values were corrected for multiple testing by the effective number of independent marker loci (6.21). +786C > T (Leu262Leu) of IL8RB showed marginal association with the peripheral blood eosinophil counts (%) among asthma patients, e.g., higher eosinophil counts (5.52 ± 5.60%) were measured among individuals who were bearing the C allele (C/C and C/T) than among those who were homozygous for the minor allele (T/T) (4.33 ± 4.09%; P c = 0.03, not significant after correction for multiple testing; Table 3). Similarly, IL8RA-B ht2 was also significantly associated with the peripheral blood eosinophil counts (%), e.g., individuals who were carrying ht2/ht2 showed significantly decreased peripheral blood eosinophil counts (3.55 ± 3.39%) compared with that of others in the study (−/− and ht2/−; 5.52 ± 5.55%; P corr = 0.018; Table 3).
Discussion
Asthma is a common and heterogeneous respiratory disease characterized by reversible airway obstruction caused by chronic inflammation of the airways. The development of asthma is determined by the interaction between host genetic susceptibility and a variety of environmental exposures (Ahmadi and Goldstein 2002; Burrows et al. 1989; Kim et al. 1999; Koh et al. 2000). The airway inflammation and remodeling are the main causes of bronchial hyperresponsiveness (BHR), which characterizes the pathophysiology of asthma. Eosinophils play an important role as effector cells in allergic inflammation (Hamid 2004).
The predominant inflammatory cell in asthmatic inflammation is the eosinophil, accompanied by T-lymphocytes, mast cells, and basophils (Denburg 1996). Eosinophilic inflammation of the airways, with an increase in activated and degranulated eosinophils is the key feature of both allergic and non-allergic asthma (Bousquet et al. 1990; Humbert et al. 1999). The extent of eosinophilic inflammation is a determinant of the severity of asthma symptoms (Bousquet et al. 1990). In addition, the number of peripheral blood eosinophils is usually elevated in both atopic and non-atopic asthmatics and is correlated with airflow limitation (Oehling et al. 1992). IL8 has been reported to be a potent eosinophil chemoattractant and stimulus for degranulation (Djukanovic 2000), and IL8 mediates chemoattraction via IL8RA and IL8RB. Based on these properties, IL8RA and IL8RB are possibly associated with the release of eosinophils, which is significantly correlated with asthma symptoms (Humbert 1996). Whereas the cooperative actions of IL5 and eotaxin are also evident in the growth, differentiation, priming, and survival of eosinophils (Gauvreau et al. 1999; Oehling et al. 1992), CXC chemokines such as IL8 play a biologic role in eosinophil activation, particularly in allergic diseases. Eosinophils from atopic patients respond to IL8, whereas eosinophils from non-atopic individuals are generally unresponsive. IL8 causes an increase in intracellular calcium concentrations, actin polymerization, and chemotaxis in IL5-primed eosinophils (Schweizer et al. 1994; Warringa et al. 1991). In animal experiments, intradermal injection of IL8 has been shown to cause a dose-dependent eosinophil accumulation (Collins et al. 1993).
The role of IL8 polymorphisms in chronic inflammatory diseases such as asthma has been shown in several studies (Heinzmann et al. 2004; Hull et al. 2000, 2001). In their studies with European participants, the −251T and 781C alleles were significantly associated with development of asthma. However, these polymorphisms were not detected in our study with a Korean population, which is possibly due to the different ethnic genetic background of the participants.
Among four nonsynonymous amino acid substitutions in IL8RA (+92T > G [M31R], +827G > C [S276T], +836G > A [R279H] and +1003C > T [R335C]), two substitutions (S276T and R279H) are located on seven transmembrane receptor domains (Liu et al. 2005).
In this study, we present an association of one synonymous variation in IL8RB and a positive association of a haplotype in the IL8RA-B gene cluster with peripheral blood eosinophil counts (%). The +786C > T was in an absolute LD with −8939C > T of IL8RB in which a potential allelic difference in cis-acting regulatory function of transcription was predicted by reference to the TRANSFAC database (Heinemeyer et al. 1998). The sequence surrounding the −8939C (CCCACAGT, where the third nucleotide is polymorphic) was predicted to be 100 and 86% similar to the core sequence of the AML3 (acute myeloid leukemia 3)-binding site, whereas the −8939T allele (CCTACAGT) completely lost both similarities. In addition, it is worth noting the weak effects of IL8 polymorphisms (IL8 −352T > A and IL8 +2670A > T) on eosinophil counts (%), although they are not statistically significant (P c = 0.07; Table 3).
In summary, we have identified 20 polymorphic sites in IL8, IL8RA, and IL8RB (including eight novel SNPs) and examined their genetic association with asthma and its related phenotypes in a Korean population (n = 1,439). Our study suggests that polymorphisms and haplotypes in IL8RA and IL8RB might be among the genetic factors for controlling peripheral blood eosinophil counts in asthma patients. Further biological and/or functional evidence would be needed to confirm the association of IL8RA and IL8RB polymorphisms in this study.
References
Ahmadi KR, Goldstein DB (2002) Multifactorial diseases: asthma genetics point the way. Curr Biol 12:R702–R704
Amin K, Ludviksdottir D, Janson C, Nettelbladt O, Bjornsson E, Roomans GM, Boman G, Seveus L, Venge P (2000) Inflammation and structural changes in the airways of patients with atopic and nonatopic asthma. BHR Group. Am J Respir Crit Care Med 162:2295–2301
Bentley AM, Menz G, Storz C, Robinson DS, Bradley B, Jeffery PK, Durham SR, Kay AB (1992) Identification of T lymphocytes, macrophages, and activated eosinophils in the bronchial mucosa in intrinsic asthma. Relationship to symptoms and bronchial responsiveness. Am Rev Respir Dis 146:500–506
Bousquet J, Chanez P, Lacoste JY, Barneon G, Ghavanian N, Enander I, Venge P, Ahlstedt S, Simony-Lafontaine J, Godard P et al (1990) Eosinophilic inflammation in asthma. N Engl J Med 323:1033–1039
Broide DH, Lotz M, Cuomo AJ, Coburn DA, Federman EC, Wasserman SI (1992) Cytokines in symptomatic asthma airways. J Allergy Clin Immunol 89:958–967
Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG (1989) Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med 320:271–277
Chung KF, Barnes PJ (1999) Cytokines in asthma. Thorax 54:825–857
Collins PD, Weg VB, Faccioli LH, Watson ML, Moqbel R, Williams TJ (1993) Eosinophil accumulation induced by human interleukin-8 in the guinea-pig in vivo. Immunology 79:312–318
Denburg JA (1996) The inflammatory response. Am J Respir Crit Care Med 153:S11–S13
Desreumaux P, Capron M (1996) Eosinophils in allergic reactions. Curr Opin Immunol 8:790–795
Djukanovic R (2000) Asthma: a disease of inflammation and repair. J Allergy Clin Immunol 105:S522–S526
Gauvreau GM, Watson RM, O’Byrne PM (1999) Kinetics of allergen-induced airway eosinophilic cytokine production and airway inflammation. Am J Respir Crit Care Med 160:640–647
Hamid Q (2004) Eosinophils in allergic inflammation. J Allergy Clin Immunol 113:182–184
Harada A, Sekido N, Akahoshi T, Wada T, Mukaida N, Matsushima K (1994) Essential involvement of interleukin-8 (IL-8) in acute inflammation. J Leukoc Biol 56:559–564
Hedrick PW (1987) Gametic disequilibrium measures: proceed with caution. Genetics 117:331–341
Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26:362–367
Heinzmann A, Ahlert I, Kurz T, Berner R, Deichmann KA (2004) Association study suggests opposite effects of polymorphisms within IL8 on bronchial asthma and respiratory syncytial virus bronchiolitis. J Allergy Clin Immunol 114:671–676
Hull J, Thomson A, Kwiatkowski D (2000) Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax 55:1023–1027
Hull J, Ackerman H, Isles K, Usen S, Pinder M, Thomson A, Kwiatkowski D (2001) Unusual haplotypic structure of IL8, a susceptibility locus for a common respiratory virus. Am J Hum Genet 69:413–419
Humbert M (1996) Pro-eosinophilic cytokines in asthma. Clin Exp Allergy 26:123–127
Humbert M, Menz G, Ying S, Corrigan CJ, Robinson DS, Durham SR, Kay AB (1999) The immunopathology of extrinsic (atopic) and intrinsic (non-atopic) asthma: more similarities than differences. Immunol Today 20:528–533
Kim YK, Cho SH, Koh YY, Son JW, Jee YK, Lee MH, Min KU, Kim YY (1999) Skin reactivity to inhalant allergens, total serum IgE levels, and bronchial responsiveness to methacholine are increased in parents of nonatopic asthmatic children. J Allergy Clin Immunol 104:311–316
Koh YY, Jeong JH, Kim CK, Kim YK, Jee YK, Cho SH, Min KU, Kim YY (2000) Atopic status and level of bronchial responsiveness in parents of children with acute bronchiolitis. J Asthma 37:709–717
Kurashima K, Mukaida N, Fujimura M, Schroder JM, Matsuda T, Matsushima K (1996) Increase of chemokine levels in sputum precedes exacerbation of acute asthma attacks. J Leukoc Biol 59:313–316
Lee J, Horuk R, Rice GC, Bennett GL, Camerato T, Wood WI (1992) Characterization of two high affinity human interleukin-8 receptors. J Biol Chem 267:16283–16287
Liu Y, Yang S, Lin AA, Cavalli-Sforza LL, Su B (2005) Molecular evolution of CXCR1, a G protein-coupled receptor involved in signal transduction of neutrophils. J Mol Evol 61:691–696
Livak KJ (1999) Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 14:143–149
Nyholt DR (2004) A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74:765–769
Oehling AG Jr, Walker C, Virchow JC, Blaser K (1992) Correlation between blood eosinophils, T-helper cell activity markers and pulmonary function in patients with allergic and intrinsic asthma. J Invest Allergol Clin Immunol 2:295–299
Palmer LJ, Celedon JC, Chapman HA, Speizer FE, Weiss ST, Silverman EK (2003) Genome-wide linkage analysis of bronchodilator responsiveness and post-bronchodilator spirometric phenotypes in chronic obstructive pulmonary disease. Hum Mol Genet 12:1199–1210
Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA (2002) Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70:425–434
Schweizer RC, Welmers BA, Raaijmakers JA, Zanen P, Lammers JW, Koenderman L (1994) RANTES- and interleukin-8-induced responses in normal human eosinophils: effects of priming with interleukin-5. Blood 83:3697–3704
Stemmler S, Arinir U, Klein W, Rohde G, Hoffjan S, Wirkus N, Reinitz-Rademacher K, Bufe A, Schultze-Werninghaus G, Epplen JT (2005) Association of interleukin-8 receptor alpha polymorphisms with chronic obstructive pulmonary disease and asthma. Genes Immun 6:225–230
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Warringa RA, Koenderman L, Kok PT, Kreukniet J, Bruijnzeel PL (1991) Modulation and induction of eosinophil chemotaxis by granulocyte-macrophage colony-stimulating factor and interleukin-3. Blood 77:2694–2700
Xu J, Postma DS, Howard TD, Koppelman GH, Zheng SL, Stine OC, Bleecker ER, Meyers DA (2000) Major genes regulating total serum immunoglobulin E levels in families with asthma. Am J Hum Genet 67:1163–1173
Acknowledgements
This work was supported by grant number M1-0302-00-0073 of the National Research Laboratory Program as part of the National Research and Development Program from the Ministry of Science and Technology of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Hyun Sub Cheong and Hyoung Doo Shin contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Cheong, H.S., Shin, H.D., Lee, S.O. et al. Polymorphisms in interleukin 8 and its receptors (IL8, IL8RA and IL8RB) and association of common IL8 receptor variants with peripheral blood eosinophil counts. J Hum Genet 51, 781–787 (2006). https://doi.org/10.1007/s10038-006-0021-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-006-0021-5
Keywords
This article is cited by
-
IL8 gene as modifier of cystic fibrosis: unraveling the factors which influence clinical variability
Human Genetics (2016)
-
CXCR1 as a novel target for directing reactive T cells toward melanoma: implications for adoptive cell transfer immunotherapy
Cancer Immunology, Immunotherapy (2012)