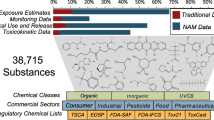
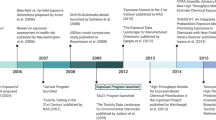
Abstract
The emerging field of computational toxicology applies mathematical and computer models and molecular biological and chemical approaches to explore both qualitative and quantitative relationships between sources of environmental pollutant exposure and adverse health outcomes. The integration of modern computing with molecular biology and chemistry will allow scientists to better prioritize data, inform decision makers on chemical risk assessments and understand a chemical's progression from the environment to the target tissue within an organism and ultimately to the key steps that trigger an adverse health effect. In this paper, several of the major research activities being sponsored by Environmental Protection Agency's National Center for Computational Toxicology are highlighted. Potential links between research in computational toxicology and human exposure science are identified. As with the traditional approaches for toxicity testing and hazard assessment, exposure science is required to inform design and interpretation of high-throughput assays. In addition, common themes inherent throughout National Center for Computational Toxicology research activities are highlighted for emphasis as exposure science advances into the 21st century.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Ankley, et al. Endocrine disrupting chemicals in fish: developing exposure indicators and predictive models of effects based on mechanism of action. submitted.
Collins F.S., Gray G.M., and Bucher J.R. Transforming environmental health protection. Science 2008: 319: 906–907.
Dix D.J., Houck K.A., Martin M.T., Richard A.M., Setzer R.W., and Kavlock R.J. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci 2007: 95 (1): 5–12.
Edwards S.W., and Preston R.J. Systems biology and mode of action based risk assessment. Toxicol Sci 2008 doi:10.1093/toxsci/kfn190.
Ekman D.R., Teng Q., et al. NMR analysis of male fathead minnow urinary metabolites: a potential approach for studying impacts of chemical exposures. Aquat Toxicol 2007: 85 (2): 104–112.
Ekman D.R., Teng Q., et al. Investigating compensation and recovery of fathead minnow (Pimephales promelas) exposed to 17alpha-ethynylestradiol with metabolite profiling. Environ Sci Technol 2008: 42 (11): 4188–4194.
Heidenfelder B.L., Reif D.M., Cohen Hubal E.A., Hudgens E.E., Bramble L.A., Wagner J.G., Harkema J.R., Morishita M., Keeler G.J., Edwards S.W., and Gallagher J.E. Comparative microarray analysis and pulmonary morphometric changes in brown Norway rats exposed to ovalbumin and/or concentrated air particulates. Submitted.
Judson R., Richard A., Dix D., Houck K., Elloumi F., Martin M., Cathey T., Transue T.R., Spencer R., and Wolf M. ACToR — aggregated computational toxicology resource. Toxicol Appl Pharmacol, Available online 18 July 2008.
Kavlock R.J., Ankley G., Blancato J., Breen M., Conolly R., Dix D., Houck K., Hubal E., Judson R., Rabinowitz J., Richard A., Setzer R.W., Shah I., Villeneuve D., and Weber E. Computational toxicology a state of the science mini review. Toxicol Sci 2007: 103 (1): 14–27.
Knudsen T.B., and Kavlock R.J. Comparative Bioinformatics and Computational Toxicology, Abbot B., and Hansen D. (Eds.). 3rd edn. Taylor & Francis 2009.
Loscalzo J., Kohane I., and Barabasi A.L. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol 2007: 3: 124.
National Research Council of the National Academies (NRC). Toxicity Testing in the 21st Century: A Vision and A Strategy. The National Academies Press, Washington, DC, 2007.
National Research Council of the National Academies (NRC). The National Children's Study Research Plan: A Review. The National Academies Press, Washington, DC, 2008.
Richard A., Yang C., and Judson R. Toxicity data informatics: supporting a new paradigm for toxicity prediction. Toxicol Mech Meth 2008: 18: 103–118.
Richard A.M., Gold L.S., and Nicklaus M.C. Chemical structure indexing of toxicity data on the Internet: Moving toward a flat world. Curr Opin Drug Discov Devel 2006: 9 (3): 314–325.
Reif D.M., et al. Integrating demographic, clinical, and environmental exposure information to identify genomic biomarkers associated with subtypes of childhood asthma. 2008 Joint Annual Conference ISEE/ISEA. Pasadena, CA.
Reif D.M., Motsinger A.A., McKinney B.A., Edwards K.M., Chanock S.J., Rock M.T., Crowe Jr J.E., and Moore J.H. Integrated analysis of genetic and proteomic data identifies biomarkers associated with systemic adverse events following smallpox vaccination. Genes Immun, published online 16 October 2008. doi:10.1038/gene.2008.80.
Schadt E.E., and Lum P.Y. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Reverse engineering gene networks to identify key drivers of complex disease phenotypes. J Lipid Res 2006: 47: 2601–2613.
Sen B., Mahadevan B., and DeMarini D.M. Transcriptional responses to complex mixtures — a review. Mutat Res 2007: 636 (2007): 144–177.
Suter G.W. Developing conceptual models for complex ecological risk assessments. Hum Ecol Risk Assess 1999: 5 (2): 375–396.
U.S. EPA. A Framework for a Computational Toxicology Research Program. Office of Research and Development, Washington, DC, 2003 EPA 600/R-03/065 http://www.epa.gov/comptox/publications/comptoxframework06_02_04.pdf.
U.S. EPA. A Pilot Study of Children's Total Exposure to Persistent Pesticides and Other Persistent Organic Pollutants (CTEPP). Volume I: Final Report. Contract Number 68-D-99-011, U.S. Environmental Protection Agency, Office of Research and Development, Research Triangle Park, NC, 2006 Available online at http://www.epa.gov/heasd/ctepp/ctepp_report.pdf.
U.S. EPA. EPA Community of Practice: Exposure Science for Toxicity Testing, Screening, and Prioritization 2008: http://www.epa.gov/ncct/practice_community/exposure_science.htmlAccessed September 16, 2008.
Weis B.K., Balshaw D., Barr J.R., Brown D., Ellisman M., Lioy P., et al. Personalized exposure assessment: promising approaches for human environmental health research. Environ Health Perspect 2005: 113 (7): 840–848.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclaimer
The US Environmental Protection Agency, through its Office of Research and Development funded and managed the research described here. It has been subjected to Agency's administrative review and approved for publication.
Rights and permissions
About this article
Cite this article
Cohen Hubal, E., Richard, A., Shah, I. et al. Exposure science and the U.S. EPA National Center for Computational Toxicology. J Expo Sci Environ Epidemiol 20, 231–236 (2010). https://doi.org/10.1038/jes.2008.70
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jes.2008.70
Keywords
This article is cited by
-
Exposure forecasting – ExpoCast – for data-poor chemicals in commerce and the environment
Journal of Exposure Science & Environmental Epidemiology (2022)
-
Integrating tools for non-targeted analysis research and chemical safety evaluations at the US EPA
Journal of Exposure Science & Environmental Epidemiology (2018)
-
Non-invasive saliva human biomonitoring: development of an in vitro platform
Journal of Exposure Science & Environmental Epidemiology (2017)
-
Internal exposure dynamics drive the Adverse Outcome Pathways of synthetic glucocorticoids in fish
Scientific Reports (2016)
-
Air pollution and health: bridging the gap from sources to health outcomes: conference summary
Air Quality, Atmosphere & Health (2012)