Abstract
We characterized in vitro activities of α-methoxyimino acylides against macrolide-resistant clinical isolates of Streptococcus pneumoniae, Streptococcus pyogenes and Mycoplasma pneumoniae with ribosome modification or substitution and selected acylide-resistant mutants to clarify the binding point of the acylides. The acylides had low MICs against erm(B) gene-containing S. pneumoniae and S. pyogenes (MIC90s, 1–4 μg ml−1). For M. pneumoniae, although they had poor potencies against macrolide-resistant strains with the A2058G (Escherichia coli numbering) mutation in 23S rRNA (MICs, >32 μg ml−1), one of them showed in vitro activities against macrolide-resistant strains with the A2058U or A2059G mutations (MICs, 0.5–1 μg ml−1). These A2058U and A2059G mutant strains were used for the selection of acylide-resistant mutants. A genetic analysis showed that new point mutations in acylide-resistant mutants were found at G2576 in domain V of 23S rRNA and at Lys90 in L22 ribosomal protein. Furthermore, a molecular modeling study revealed that G2505/C2610, which enables stacking with G2576, might interact with a pyridyl moiety or an α-methoxyimino group at the 3-position of acylides. The α-methoxyimino acylides were shown to possess a tertiary binding point at G2505/C2610 in 23S rRNA. Our results suggest that α-methoxyimino acylides represent significant progress in macrolide antimicrobials.
Similar content being viewed by others
Introduction
Macrolide antimicrobials bind to the large ribosome subunit and inhibit protein synthesis by blocking the progression of the nascent peptide in the exit tunnel. These antimicrobials, such as clarithromycin and azithromycin (Figure 1), have played a key role in the treatment of community-acquired respiratory tract infections (RTIs). However, macrolide-resistant Streptococcus pneumoniae is now a major clinical problem in some countries, especially in Europe and the Asia-Pacific region.1, 2, 3 Although Streptococcus pyogenes remains universally β-lactam susceptible, macrolide resistance has been recognized, especially in Europe and Asia.4, 5, 6, 7 Macrolide-resistant Mycoplasma pneumoniae is also emerging in pediatric populations in Japan and China.8, 9, 10 In S. pneumoniae and S. pyogenes, two major mechanisms of macrolide resistance have been reported. One is methylation of the target site nucleotide A2058 (Escherichia coli numbering used throughout) in 23S rRNA mediated by erm genes, and the other is the alteration of antibiotic accumulation as a result of mef genes.11 M. pneumoniae resistance to macrolides is caused by point mutations in domain V of the 23S rRNA gene that interfere with the binding of macrolides to rRNA.8
Ketolides are semisynthetic derivatives from erythromycin that have a keto group at the C-3 position of the lactone ring. In addition, telithromycin, a ketolide, has an alkyl-aryl extension from the cyclic carbamate at positions 11 and 12 of the lactone ring (Figure 1). The extension engages in stacking with A752 and U2609 in domain II in 23S rRNA as secondary binding point, increasing the binding affinity to bacterial ribosome.12, 13, 14 Consequently, telithromycin is active against macrolide-resistant S. pneumoniae.15, 16 However, the emergence of telithromycin-resistant clinical strains of S. pneumoniae has also been reported.17, 18 In addition, telithromycin has been reported to be active against macrolide-resistant S. pyogenes with the efflux gene mef(A) and the inducible methylase gene erm(A), but not active against most erm(B) gene-carrying S. pyogenes.19, 20, 21 Moreover, mutations in ribosomal proteins L4 and L22 reportedly confer reduced susceptibility to macrolides and/or telithromycin.17, 22, 23 Not only clarithromycin and azithromycin, but also telithromycin showed weak activities against macrolide-resistant M. pneumoniae.8 In this situation, the need for new macrolide agents to treat drug-resistance bacteria is increasingly important. Thus, many investigational macrolides have been reported across the globe.24, 25
Acylides, 3-O-acyl-erythromycin derivatives with potent activities against mef-mediated efflux and inducibly erm-containing resistant strains, were first reported by us.26, 27, 28 In addition, we have shown that TP0020827 (TP-C, formerly FMA0199, Figure 1) in which an alkyl-aryl side chain is attached to the cyclic carbamate at positions 11 and 12 of the 3-O-(2-pyridyl) acetyl derivative improve activities against erm-containing resistant S. pneumoniae.24, 29, 30
We decided to further optimize the acyl group with improved activities against resistant strains. We tried to change the conformation and electron density of the pyridine ring at the C-3 position which might have any interactions to 23S rRNA by introducing some substituents to the methylene carbon. Especially, introduction of methoxyimino group was expected to fix pyridine ring as the result of extending conjugation system from carboxylate to aromatic ring. We obtained acylide derivatives, TP0097302 (TP-B) and TP0083177 (TP-D), possessing a methoxyimino group at the α-position on the 3-O-acyl side chain (Figure 1).31 TP-D also carries the same C-11,12 extended carbamate side chain as TP-C. On the other hand, unlike ketolides, minimal research using microbiological and genetic approaches has been done to investigate the binding mode of acylides.
This paper describes the in vitro activities of α-methoxyimino acylides, TP-B and TP-D, and their (des)-metoxyimino derivatives, TP0017383 (TP-A) and TP-C, against macrolide-resistant clinical isolates caused by ribosome modification or substitution, including M. pneumoniae. We also discuss the binding points of acylides in ribosome based on the results of selection and analysis of acylide-resistant mutants from macrolide-resistant clinical strains of M. pneumoniae.
Materials and methods
Compounds
TP0017383 (TP-A),32 TP0097302 (TP-B),31 TP0020827 (TP-C),29, 30 TP0083177 (TP-D)31 and TP0020828 (TP-E),33 a ketolide, as a (des)-3-O-acyl derivative (Figure 1) were chemically synthesized by Taisho Pharmaceuticals Co., Ltd (Saitama, Japan). Azithromycin was purchased from U.S. Pharmacopeial Convention (Rockville, MD, USA) and Sigma-Aldrich (St. Louis, MO, USA). Clarithromycin, clindamycin and minocycline were purchased from Sigma-Aldrich.
Bacterial strains
S. pneumoniae ATCC 49619, ATCC 700904 and M. pneumoniae ATCC 15531 were purchased from the American Type Culture Collection. Most of the clinical isolates used in the MIC determination study and to select for acylides-resistant mutants were obtained in Japan. These strains of streptococci and M. pneumoniae were collected during 2006–2009 and in 2011, respectively. The erm(B) genes in streptococci were detected by PCR amplification using a resistant gene detection kit obtained from Wakunaga Pharmaceutical Co., Ltd (Hiroshima, Japan).
MIC determination
The MICs of each compound for streptococci were determined using the broth microdilution method, according to the Clinical and Laboratory Standards Institute guidelines.34, 35
The MICs for M. pneumoniae were determined using a broth microdilution method.36 PPLO broth (Difco Inc., Detroit, MI, USA) containing 20% horse serum, 2.5% yeast extract, 1% glucose (Sigma-Aldrich) and 0.002% phenol red (Sigma-Aldrich) was used as the growth medium. A suspension estimated to be 105 CFU ml−1 was added to each well, which contained 100 μl of the medium. The microplate was incubated aerobically over 5 days at 37 °C under moist conditions until a color change in the antibiotic-free growth control was confirmed. The MIC was defined as the lowest concentration of each antibiotic without a color change.
Selection of acylide-resistant mutants of M. pneumoniae strains
The selection of acylide-resistant mutants was performed by serial transfers of M. pneumoniae strains in PPLO broth containing 2, 8 or 32 times the MIC of each acylide. For the first passage, the culture was incubated aerobically for 10–31 days at 37 °C until a color change was confirmed. The isolates were stored at −70 °C. For the second passage, the culture was incubated aerobically for 6–12 days at 37 °C until a color change was confirmed. After all the isolates were grown in antibiotic-free medium, the MICs of the acylides and reference compounds were determined.
PCR amplification and DNA sequencing
The total length of the 23S rRNA gene in the parent and selected strains of M. pneumoniae was sequenced. Two primer sets (5′-CAATCCGTTACTAAGGGCTTATGGT-3′ and 5′-TCCAATAAGTCCTCGAGCAATTA-3′) were used for amplification of the entire 23S rRNA gene. A 0.5-ml growth culture of each M. pneumoniae strain was centrifuged at 13 000 r.p.m. for 10 min at 4 °C. After removal of the supernatant, the sediment was suspended in 30 μl of Gene Releaser (Funakoshi, Tokyo, Japan) and boiled for 10 min. PCRs were performed using a TaKaRa-Bio thermal cycler (TP 3100) with two primers, DyNazyme EXT DNA polymerase (Thermo Scientific, Waltham, MA, USA) and a template. Amplification was achieved with an initial denaturation step of 1 min at 94 °C, 35 cycles of 20 s at 94 °C, 30 s at 53 °C for annealing and a 3-min extension step at 72 °C for an ~2900-bp fragment of 23S rRNA. After purification with the QIAquick PCR purification kit (QIAGEN, Hilden, Germany), both strands of the PCR products were directly sequenced with a BigDye Terminator v3.1 Cycle Sequencing Kit and an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions. Specific oligonucleotidic primers for regions of interest in genes encoding 23S rRNA were designed from the complete genome sequence.37 The results were compared with the parent strains and the selected mutants.
Genes encoding ribosomal proteins L4 and L22 were sequenced using primers described by Pereyre et al.38 and Matsuoka et al.39
Molecular modeling studies
All the acylides used in this study were composed of a 14-membered macrolactone ring with a desosamine sugar at position 5 and a carbamate heterocycle involving the carbon atoms at positions 11 and 12. Furthermore, TP-C and TP-D contain an alkyl-aryl arm attached to a carbamate heterocycle. These chemical characteristics are similar to those of the ketolide telithromycin. Therefore, the X-ray crystallographic structure of E. coli ribosome bound to telithromycin (Protein Data Bank: 4V7S)14 was used as a template structure in this study.
Molecular docking studies were conducted using molecular operating environment (MOE; version MOE 2014.09; Chemical Computing Group Inc., Quebec, Canada). As working on the complete structure of the E. coli ribosome bound to telithromycin is computationally demanding, a partial structure was built. Ribosomal protein amino acids and rRNA nucleobases residing within 20 Å of the telithromycin were extracted using the MOE package. The structure of the acylide TP-D was drawn in the MOE package by replacing the ribosome-bound telithromycin atoms corresponding to the 14-membered macrolactone ring, the desosamine sugar, the carbamate heterocycle and the alkyl-aryl arm attached to the carbamate heterocycle. The pyridyl acetyl group at position 3 of TP-D was drawn toward G2505/C2610 of the 23S rRNA. To obtain the docking pose, TP-D and the partial ribosome were prepared with MOE by adding hydrogen atoms, assigning a partial atomic charges force field, and by performing an energy minimization of the pyridyl ring of the alkyl-aryl arm and the pyridyl acetyl group at position 3 of TP-D with the other atoms fixed at their position to an RMS gradient of 0.1 kcal/mol/Å2.
Results
Antibacterial activities of acylides against streptococci
The MICs of four acylides were first determined for the ATCC reference strains (Table 1). All the acylides and TP-E, a ketolide with the same side chain at the 11, 12-position as TP-C and α-methoxyimino acylide TP-D, showed excellent in vitro activities against S. pneumoniae ATCC 49619, a macrolide-susceptible strain. The activity of TP-D (MIC: 0.25 μg ml−1) was slightly inferior to those of the other acylides (MIC: 0.015 to 0.12 μg ml−1). In contrast, the S. pneumoniae ATCC 700904 strain is a macrolide-resistant strain with the erm(B) gene.40 Clarithromycin, azithromycin and clindamycin had MICs of >128 μg ml−1. The plain-type acylide TP-A and the ketolide TP-E were weakly active against ATCC 700904 (MICs: 64 and 16 μg ml−1, respectively). Interestingly, α-methoxyimino acylide TP-B exhibited an improved antibacterial activity against ATCC 700904. In addition, TP-C, which has a side chain at the 11, 12-position, also exhibited an improved antibacterial activity. TP-D had the best activity against ATCC 700904 (MIC: 0.5 μg ml−1).
The MIC ranges, MIC50 and MIC90, of the four acylides and reference compounds against Japanese clinical isolates of erm(B) gene-carrying S. pneumoniae and S. pyogenes are summarized in Table 2. All the erm(B) gene-carrying strains of both streptococci were strongly resistant to clarithromycin, azithromycin and clindamycin, with MICs of 128 or >128 μg ml−1. The plain-type acylide TP-A was weakly active against both erm(B) gene-carrying S. pneumoniae (MIC range: 16–128 μg ml−1) and S. pyogenes (MIC range: 128 to >128 μg ml−1). However, three acylides, TP-B, TP-C and TP-D, had MICs of ⩽8 μg ml−1 against both streptococci. In addition, the MIC90s of TP-B, TP-C and TP-D against S. pneumoniae were 4, 2 and 1 μg ml−1, respectively, and these values were comparable to those against S. pyogenes. In particular, TP-D was active against streptococci over a narrow range (0.5–1 μg ml−1). Although TP-E had MIC50 and MIC90 values of 1 and 64 μg ml−1 against S. pneumoniae, respectively, the ketolide had MICs of >128 μg ml−1 against almost all the erm(B) gene-carrying S. pyogenes. Therefore, the activity of TP-D was 128-fold or more potent than that of ketolide TP-E against erm(B) gene-carrying S. pyogenes.
Antibacterial activities of acylides against M. pneumoniae
The MICs of the acylides against macrolide-resistant M. pneumoniae are shown in Table 3. Although all clinical isolates were resistant to a 14-membered macrolide, the 15-memberd macrolide azithromycin was weakly active against A2058U and A2059G mutants, compared with its effect against an A2058G mutant, as previously reported.39, 41, 42 Three acylides that were active against macrolide-resistant streptococci were used. The MICs of the three acylides were 0.001–0.008 μg ml−1 against M. pneumoniae ATCC 15311, a macrolide-susceptible strain, and these values were comparable to that of clarithromycin. Furthermore, TP-C and TP-D showed antibacterial activities with MICs of 0.25–1 μg ml−1 against strains of macrolide-resistant M. pneumoniae with A2058U or A2059G mutations in 23S rRNA. However, the two acylides were not active at 64 or >64 μg ml−1 against macrolide-resistant strains with the A2058G mutation in 23S rRNA. In addition, TP-B was not active at >32 μg ml−1 against all the macrolide-resistant strains that were tested.
Selection of acylides-resistant mutants of M. pneumoniae
To assess the mechanism and binding mode of action of acylides for bacterial ribosome, the selection of mutants resistant to acylides was conducted by two passages of M. pneumoniae 6869, 6941 and 6937 as the parental strains at 2-, 8- or 32-fold the MIC of each acylide. Eleven acylide-resistant mutants were selected: five mutants from parent strain 6869, four mutants from parent strain 6941 and two mutants from parent strain 6937 (Table 4). On the other hand, mutants were not selected at 32-fold the MIC of TP-C from parent strain 6869, at 32-fold the MIC from parent strain 6941, or at 32- and 8-fold the MIC for parent strain 6937. Some of the mutants showed significantly increased resistance to the reference macrolides, particularly azithromycin. Furthermore, four high-level resistant mutants to each acylide (⩾16 μg ml−1) were obtained with both selector acylides from parent strains 6869 and 6937.
Analysis of 23S rRNA and L4 and L22 sequences in mutants
Among the nine mutants from parent strains with the A2058U mutation, one of the high-level resistant mutants selected from passage in TP-D had an A2058C substitution, one selected from passage in TP-C had a G2057A mutation in addition to an original A2058U mutation, three selected from passage in each acylide had a G2576U mutation in addition to an original A2058U mutation, two mutants had both A2058U and C2611U mutations, and two mutants had not only an original A2058U mutation in domain V of 23S rRNA but also a Lys90Asn mutation in ribosomal protein L22 (Table 4). Each mutant with the same double mutations resulted in the same phenotype of resistance. For mutants with both A2058U and G2576U mutations, the reference macrolides were less effective. However, for mutants with both A2058U and Lys90Asn, the MICs of the reference macrolides were unchanged.
Two mutants selected from a parent strain with the A2059G mutation harbored not only an original A2059G mutation, but also a Lys90Glu mutation in ribosomal protein L22 (Table 4). The MICs of all the compounds were significantly higher against the mutants.
None of the mutants exhibited changes in ribosomal protein L4 compared with the parent strains.
Molecular modeling studies
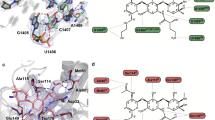
The result of the molecular modeling studies for TP-D is shown in Figure 2. The analysis indicated that a pyridyl group or an α-methoxyimino group at the 3-position of TP-D has an interaction with G2505/C2610 on 23S rRNA (Figure 2a). Meanwhile, the pyridyl group in the side chain at the 11, 12-position of TP-D was determined not only to have a stacking interaction with A752-U2609 base pair on 23S rRNA but also to position near Lys90 in L22 (Figure 2b).
Stereo view of docked poses of TP-D in the peptidyl transferase center of 23S rRNA. (a) TP-D (yellow carbon balls) and G2576 (light blue carbon balls) are represented by ball and stick models. Other nucleobases (orange carbon sticks) are represented by stick models. (b) TP-D (yellow carbon balls) and Lys90 (light blue carbon balls) are represented by ball and stick models. Other nucleobases (orange carbon sticks) and ribosomal protein L22 are represented by stick and ribbon models, respectively.
Discussion
The tested ketolide TP-E was not active against most erm(B) gene-containing S. pyogenes strains, but was partly active against erm(B) gene-containing S. pneumoniae. Nevertheless, the acylide TP-B with a methoxyimino group at the α-position on the 3-O-acyl side chain showed potent antibacterial activities against erm(B) gene-containing strains of both S. pneumoniae and S. pyogenes (Table 2). In addition, the in vitro activity of TP-B against erm(B) gene-carrying S. pneumoniae and S. pyogenes was superior than that of TP-A (Table 2). In S. pyogenes, clindamycin-resistant erm(B) gene-containing strains are usually constitutive resistant phenotype.43 Therefore, we realize that the erm(B) in the tested S. pyogenes strains would be constitutive. Increased activity of the α-methoxyimino acylides against erm(B) gene-containing strains could not be explained by their inability to sufficiently induce the expression of the methylase. Thus, we expected that the α-methoxyimino acylides possess other biding point in 23S rRNA to improve antibacterial activity.
To select for acylide-resistant mutants and to promote a better understanding of their binding contribution, we propagated in vitro macrolide-resistant but acylide-susceptible M. pneumoniae clinical isolates in media containing acylides at concentrations above the MICs. Although macrolide-resistant clinical isolates of streptococci containing the erm(B) gene were used in this study, we expected that obtaining a relatively high-level acylide-resistant mutant with a specific secondary or tertiary binding point in a small number of rrn operons (4 or 6 rrn operons in S. pneumoniae or S. pyogenes, respectively) would be difficult.44 On the other hand, the genome of M. pneumoniae is entirely sequenced, and only a single rRNA gene operon is known to exist.37, 44 Thus, although M. pneumoniae is slow growing, we used clinical isolates of M. pneumoniae as useful tools for characterizing acylides.
We performed a multistep resistance study using TP-C or TP-D. Acylide-resistant mutants were obtained after a second passage (Table 4). A2058, G2057 and C2611 have been characterized in some bacterial species.39, 44, 45, 46 The available high-resolution crystallographic structures suggest that the central macrolactone ring of 14-membered macrolide compounds establishes a hydrophobic interaction with nucleotides 2057, 2058 and 2611 in domain V, which partly form the tunnel wall on the side of the peptidyl transferase center.47 Thus, G2057, A2058 and C2611 are unlikely to contribute to the specific interaction of acylides with 23S rRNA.
To our knowledge, the G2576U mutation in 14- and 15-membered macrolide-resistant mutants has never been reported in M. pneumoniae or any other microorganism. This transition has been described for a 16-membered macrolide josamycin-resistant in vitro selection mutant in Mycoplasma hominis.48 Interestingly, the G2576U transversion in 23S rRNA was previously described frequently in oxazolidinone class linezolid-resistant clinical isolates and in vitro linezolid-selected mutants.49, 50 Unfortunately, linezolid is inactive against M. pneumoniae.51, 52 Therefore, we could not characterize the contribution of the G2576U mutation in M. pneumoniae to linezolid resistance directly. The linezolid-binding pocket is lined by eight nucleotides, G2061, A2451, C2452, A2503, U2504, G2505, U2506 and U2585, which interact directly with linezolid in 23S rRNA.53, 54, 55 G2576 does not interact with linezolid directly, but this nucleotide is located behind the linezolid-binding pocket and stacks directly onto G2505.53, 54, 55 Long et al.56 proposed that G2576U transversion would presumably reduce the degree of direct stacking onto G2505 and disrupt the interaction with the U2506 backbone that adjoins the bound linezolid; consequently, the transversion would decrease linezolid binding. Based on the crystal structure, linezolid binds to a site near a neighboring, but not directly overlapping, the macrolide biding site in 23S rRNA.57 On the other hand, the study of X-ray co-crystal structure has shown that the cladinose at 3-position of 14 or 15-memberd macrolides comes into close contact with G2505 and C2610 in domain V.14, 58, 59 However, Magee et al.60 showed that some carbamolides, one of the C-3 substituent macrolide, formed an additional interaction with G2505 and C2610 from an X-ray co-crystal structure of the Deinococcus radiodurans 50S ribosome. Our proposed binding mode of TP-D would indicate an interaction between G2505/C2610 and a pyridyl group or an α-methoxyimino group at the 3-position (Figure 2a). Therefore, our results of mutation analysis suggest that some C-3 substituent macrolides may interact directly with G2505/C2610 and exert antibacterial activities against macrolide-resistant pathogens through a modification at its binding site on the ribosome.
Unfortunately, TP-B could not be used in the selection of acylides-resistant mutants of M. pneumoniae because of the high MICs of TP-B against any macrolide-resistant M. pneumoniae strains (Table 3). The methoxyimino group presumably improved the rigidity of the pyridyl acetyl moiety. The rigidity of the 3-O-acyl side chain of TP-B might contribute to an improved affinity to dimethylated 23S rRNA in streptococci through an interaction with G2505/C2610.
Some investigators have indicated that mutations or an insertion in ribosomal protein L22 could confer resistance to ketolides.22, 23, 61 However, Lys90Asn or Glu mutations in ribosomal protein L22 had never been reported in any macrolide/ketolide-resistant bacteria. Moreover, our docking study showed that Lys90 in L22 was located close to the pyridyl moiety of TP-D (Figure 2b). The structure of the solithromycin and ribosome complex shows that the Lys90 is located reasonably close to the alkyl-aryl side chain.62 Therefore, the pyridyl moiety of the 11, 12 side chain of TP-C and -D could also undergo a stacking interaction with the A752-U2609 base pair and/or come in contact with Lys90 in L22, similar to ketolides. In addition, based on the similar activities of TP-C and TP-D against macrolide-resistant strains (Tables 2 and 3), we speculated that the possession of both secondary and tertiary binding points in 23S rRNA could allow acylides to bind strongly to ribosomes with or without the rigidity of the 3-O-acyl side chain enabled by a methoxyimino group.
In conclusion, α-methoxyimino acylides were shown to have a tertiary binding point at G2505/C2610 in 23S rRNA. Our results suggest that α-methoxyimino acylides represent significant progress in macrolide antimicrobials. These findings provide a foundation for further optimization to improve antibacterial activities against macrolide-resistant pathogens, including M. pneumoniae.
References
Farrell, D. J., Couturier, C. & Hryniewicz, W. Distribution and antibacterial susceptibility of macrolide resistance genotypes in Streptococcus pneumoniae: PROTEKT Year 5 (2003-2004). Int. J. Antimicrob. Agents 31, 245–249 (2008).
Kim, S. H. et al. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob. Agents Chemother. 56, 1418–1426 (2012).
Farrell, D. J., Flamm, R. K., Sader, H. S. & Jones, R. N. Results from the Solithromycin International Surveillance Program (2014). Antimicrob. Agents Chemother. 60, 3662–3668 (2016).
Pérez-Trallero, E. et al. Antimicrobial resistance among respiratory pathogens in Spain: latest data and changes over 11 years (1996-1997 to 2006-2007). Antimicrob. Agents Chemother. 54, 2953–5959 (2010).
Michos, A. et al. Molecular analysis of Streptococcus pyogenes macrolide resistance of paediatric isolates during a 7 year period (2007-13). J. Antimicrob. Chemother. 71, 2113–2117 (2016).
Wajima, T. et al. Distribution of emm type and antibiotic susceptibility of group A streptococci causing invasive and noninvasive disease. J. Med. Microbiol. 57, 1383–1388 (2008).
Huang, C. Y. et al. Epidemiology and molecular characterization of macrolide-resistant Streptococcus pyogenes in Taiwan. J. Clin. Microbiol. 52, 508–516 (2014).
Morozumi, M., Takahashi, T. & Ubukata, K. Macrolide-resistant Mycoplasma pneumoniae: characteristics of isolates and clinical aspects of community-acquired pneumonia. J. Infect. Chemother. 16, 78–86 (2010).
Kawai, Y. et al. Nationwide surveillance of macrolide-resistant Mycoplasma pneumoniae infection in pediatric patients. Antimicrob. Agents Chemother. 57, 4046–4049 (2013).
Zhao, F. et al. Surveillance of macrolide-resistant Mycoplasma pneumoniae in Beijing, China, from 2008 to 2012. Antimicrob. Agents Chemother. 57, 1521–1523 (2013).
Leclercq, R. & Courvalin, P. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46, 2727–2734 (2002).
Hansen, L. H., Mauvais, P. & Douthwaite, S. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31, 623–631 (1999).
Xiong, L., Shah, S., Mauvais, P. & Mankin, A. S. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31, 633–639 (1999).
Dunkle, J. A., Xiong, L., Mankin, A. S. & Cate, J. H. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl Acad. Sci. USA 107, 17152–17157 (2010).
Leclercq, R. Overcoming antimicrobial resistance: profile of a new ketolide antibacterial, telithromycin. J. Antimicrob. Chemother. 48 (Suppl. B), 9–23 (2001).
Lonks, J. R. & Goldmann, D. A. Telithromycin: a ketolide antibiotic for treatment of respiratory tract infections. Clin. Infect. Dis. 40, 1657–1664 (2005).
Wolter, N., Smith, A. M., Low, D. E. & Klugman, K. P. High-level telithromycin resistance in a clinical isolate of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51, 1092–1095 (2007).
Wolter, N. et al. Telithromycin resistance in Streptococcus pneumoniae is conferred by a deletion in the leader sequence of erm(B) that increases rRNA methylation. Antimicrob. Agents Chemother. 52, 435–440 (2008).
Farrell, D. J., Morrissey, I., Bakker, S. & Felmingham, D. Molecular characterization of macrolide resistance mechanisms among Streptococcus pneumoniae and Streptococcus pyogenes isolated from the PROTEKT 1999-2000 study. J. Antimicrob. Chemother. 50 (Suppl. S1), 39–47 (2002).
Nagai, K. et al. Susceptibility to telithromycin in 1,011 Streptococcus pyogenes isolates from 10 central and Eastern European countries. Antimicrob. Agents Chemother. 46, 546–549 (2002).
Morosini, M. I. et al. Streptococcus pyogenes isolates with characterized macrolide resistance mechanisms in Spain: in vitro activities of telithromycin and cethromycin. J. Antimicrob. Chemother. 52, 50–55 (2003).
Canu, A. et al. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46, 125–131 (2002).
Walsh, F., Willcock, J. & Amyes, S. High-level telithromycin resistance in laboratory-generated mutants of Streptococcus pneumoniae. J. Antimicrob. Chemother. 52, 345–353 (2003).
Asaka, T., Manaka, A. & Sugiyama, H. Recent developments in macrolide antimicrobial research. Curr. Top. Med. Chem. 3, 961–989 (2003).
Liang, J. H. & Han, X. Structure-activity relationships and mechanism of action of macrolides derived from erythromycin as antibacterial agents. Curr. Top. Med. Chem. 13, 3131–3164 (2013).
Asaka, T. et al 37th Interscience Conference on Antimicrobial Agents and Chemotherapy (Abstract no. F-262, Toronto, Canada, (1997).
Tanikawa, T. et al. Synthesis and antibacterial activity of acylides (3-O-acyl-erythromycin derivatives): a novel class of macrolide antibiotics. J. Med. Chem. 44, 4027–4030 (2001).
Tanikawa, T. et al. Synthesis and antibacterial activity of a novel series of acylides: 3-O-(3-pyridyl)acetylerythromycin A derivatives. J. Med. Chem. 46, 2706–2715 (2003).
Asaka, T. et al (Taisho Pharmaceutical Co., Ltd.). Erythromycin A derivatives. WO 1998023628 A1 (1998).
Asaka, T. et al 39th Interscience Conference on Antimicrobial Agents and Chemotherapy (Abstract no. F-2159, San Francisco, CA, USA, (1999).
Asaka, T., Kashimura, M., Manaka, A., Tanikawa, T. & Sugimoto, T. (Taisho Pharmaceutical Co., Ltd.). Macrolide derivative. JP 2001072699A (2001).
Kashimura, M., Kawamura, M., Asaka, T., Takayama, K. & Ogita, H. (Taisho Pharmaceutical Co., Ltd.). Macrolide derivative. WO 2007129646 A1 (2007).
Asaka, T. et al (Taisho Pharmaceutical Co., Ltd.). Erythromycin A derivatives. WO 1998040392 A1 (1998).
Clinical and Laboratory Standards Institute Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Approved Standard M7-A09, Clinical and Laboratory Standards Institute, Wayne, PA, USA, (2012).
Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing. 22th Informational Supplement. M100-S22, Clinical and Laboratory Standards Institute, Wayne, PA, USA, (2012).
Ishida, K. et al. In vitro and in vivo activities of macrolides against Mycoplasma pneumoniae. Antimicrob. Agents Chemother. 38, 790–798 (1994).
Himmelreich, R. et al. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24, 4420–4449 (1996).
Pereyre, S. et al. In vitro selection and characterization of resistance to macrolides and related antibiotics in Mycoplasma pneumoniae. Antimicrob. Agents Chemother. 48, 460–465 (2004).
Matsuoka, M. et al. Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob. Agents Chemother. 48, 4624–4630 (2004).
McGee, L. et al. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39, 2565–2571 (2001).
Suzuki, Y. et al. Community outbreak of macrolide-resistant Mycoplasma pneumoniae in Yamagata, Japan in 2009. Pediatr. Infect. Dis. J. 32, 237–240 (2013).
Matsuda, K. et al. Gene and cytokine profile analysis of macrolide-resistant Mycoplasma pneumoniae infection in Fukuoka, Japan. BMC Infect. Dis. 13, 591 (2013).
Giovanetti, E., Montanari, M. P., Mingoia, M. & Varaldo, P. E. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43, 1935–1940 (1999).
Vester, B. & Douthwaite, S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45, 1–12 (2001).
Furneri, P. M., Rappazzo, G., Musumarra, M. P., Tempera, G. & Roccasalva, L. S. Genetic basis of natural resistance to erythromycin in Mycoplasma hominis. J. Antimicrob. Chemother. 45, 547–548 (2000).
Ross, J. I. et al. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob. Agents Chemother. 41, 1162–1165 (1997).
Kannan, K. & Mankin, A. S. Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Ann. N. Y. Acad. Sci. 1241, 33–47 (2011).
Pereyre, S., Renaudin, H., Charron, A., Bébéar, C. & Bébéar, C. M. Emergence of a 23S rRNA mutation in Mycoplasma hominis associated with a loss of the intrinsic resistance to erythromycin and azithromycin. J. Antimicrob. Chemother. 57, 753–756 (2006).
Tsiodras, S. et al. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358, 207–208 (2001).
Long, K. S. & Vester, B. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56, 603–612 (2012).
Kenny, G. E. & Cartwright, F. D. Susceptibilities of Mycoplasma hominis M. pneumoniae, and Ureaplasma urealyticum to GAR-936, dalfopristin, dirithromycin, evernimicin, gatifloxacin, linezolid, moxifloxacin, quinupristin-dalfopristin, and telithromycin compared to their susceptibilities to reference macrolides, tetracyclines, and quinolones. Antimicrob. Agents Chemother. 45, 2604–2608 (2001).
Waites, K. B., Crabb, D. M. & Duffy, L. B. Comparative in vitro susceptibilities of human mycoplasmas and ureaplasmas to a new investigational ketolide, CEM-101. Antimicrob. Agents Chemother. 53, 2139–2141 (2009).
Wilson, D. N. et al. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl Acad. Sci. USA 105, 13339–13344 (2008).
Ippolito, J. A. et al. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 51, 3353–3356 (2008).
Eyal, Z. et al. Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc. Natl Acad. Sci. USA 112, E5805–E5814 (2015).
Long, K. S. et al. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross-resistance. Antimicrob. Agents Chemother. 54, 4705–4713 (2010).
Blanchard, S. C., Cooperman, B. S. & Wilson, D. N. Probing translation with small-molecule inhibitors. Chem. Biol. 17, 633–645 (2010).
Schlünzen, F. et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413, 814–821 (2001).
Hansen, J. L. et al. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10, 117–128 (2002).
Magee, T. V. et al. Novel 3-O-carbamoyl erythromycin A derivatives (carbamolides) with activity against resistant staphylococcal and streptococcal isolates. Bioorg. Med. Chem. Lett. 23, 1727–1731 (2013).
Berisio, R., Corti, N., Pfister, P., Yonath, A. & Böttger, E. C. 23S rRNA 2058A→G alteration mediates ketolide resistance in combination with deletion in L22. Antimicrob. Agents Chemother. 50, 3816–3823 (2006).
Llano-Sotelo, B. et al. Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis. Antimicrob. Agents Chemother. 54, 4961–4970 (2010).
Acknowledgements
We thank N Fukuhara, A Sugita, H Nabeta, M Kawamura, S Murakami, T Aida and K Taniguchi for technical support with the experiments and/or productive discussions. We also thank T Sugimoto for productive discussions and supplying necessary compounds.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Sugiyama, H., Yoshida, I., Ueki, M. et al. In vitro antibacterial activity of α-methoxyimino acylide derivatives against macrolide-resistant pathogens and mutation analysis in 23S rRNA. J Antibiot 70, 264–271 (2017). https://doi.org/10.1038/ja.2016.148
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2016.148