Abstract
Kutznerides are hexadepsipeptide antifungal and antimicrobial agents containing O-methyl-L-serine in their very unique peptidic backbone. During kutznerides biosynthesis, this O-methylated amino-acid residue is proposed to result from the action of an adenylation (A) domain present in KtzH, which is interrupted by the S-adenosylmethionine-binding-containing part of a methyltransferase. In this study, we co-expressed recombinant KtzH(A4MA4T4) with its MbtH-like protein partner KtzJ and demonstrated the requirement for KtzJ in producing soluble and active KtzH(A4MA4T4). We demonstrated the specificity of KtzH(A4MA4T4) toward L-Ser and showed the activity of the partial methyltransferase enzyme in O-methylation of L-Ser after its covalent attachment to the thiolation domain of KtzH(A4MA4T4). The insights gained from this work may guide future study and development of engineered interrupted adenylation domains for combinatorial biosynthetic methodologies.
Similar content being viewed by others
Introduction
The antifungal and antimicrobial agents kutznerides are nonribosomal cyclic hexadepsipetides derived from the soil actinomycete Kutzneria sp. 744.1, 2 Nonribosomal peptides are built on large multi-modular assembly-lines termed nonribosomal peptide synthetases. Each nonribosomal peptide synthetase module comprises a minimum of 3 core domains: an adenylation (A) domain responsible for activation of an amino-acid building block to its AMP counterpart, a thiolation (T) domain onto which the activated amino-acid building block gets covalently attached and a condensation (C) domain responsible for linking the amino-acid building blocks tethered to the T domains surrounding it. Additional auxiliary domains such as methyltransferases (M) can be embedded into the assembly-line to increase structural diversity of the natural product generated.
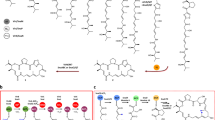
The skeleton of the unique and highly structurally diverse nonribosomally biosynthesized kutznerides is composed of an α-hydroxy acid, either (S)-2-hydroxy-3,3-dimethylbutyric acid (OHdiMeBu) or (S)-2-hydroxy-3-methylbutyric acid (OHMeBu) and five rare nonproteinogenic amino acids: D-piperazic acid (Pip), O-methyl-L-serine (O-Me-L-Ser), the erythro or threo isomer of 3-hydroxy-D-glutamic acid (3-OH-Glu), (2S,3aR,8aS)-6,7-dichloro-3a-hydroxy-hexahydropyrrolo[2,3-b]indole-2-carboxylic acid (diClPIC) and 2-(1-methylcyclopropyl)-D-glycine (D-mecPG) (Figure 1a).
(a) Structures of kutznerides 1–9 with the O-methylated L-Ser, formation of which is described in this study, marked by a box. (b) Kutznerides’ entire gene cluster. White, pale gray and dark gray arrows represent genes encoding for enzymes previously biochemically characterized, not studied and studied herein, respectively. (c) Structural organization of the kutzneride NRPS. Here, A denotes adenylation domain; C, condensation domain; E, epimerization domain; KR, ketoreductase domain; M, methyltransferase domain; T, thiolation domain (note: in the literature, T domains are also referred to as carrier protein and peptidyl carrier protein domains denoted as CP or P, PC and PCP, respectively); TE, thioesterase domain. (d) The general overview of the activity of the KtzH(A4MA4T4) and KtzJ enzyme pair studied herein.
Soon after the discovery of the kutznerides, Walsh and co-workers3 identified a biosynthetic gene cluster comprised of three potential nonribosomal peptide synthetases, KtzE, KtzG and KtzH, responsible for their formation (Figure 1b). His research team also elucidated a number of important biochemical transformations involved in the production of the unique building blocks that compose the kutznerides structural scaffold. They performed pioneering studies towards understanding Pip biosynthesis by establishing KtzI as an ornithine N-hydroxylase responsible for the conversion of ornithine into N5-hydroxy-ornithine, the first committed step during Pip formation.4 They identified KthP as the halogenase responsible for the chlorination of Pip.5 They discovered KtzO and KtzP as the nonheme FeII/α-ketoglutarate-dependent hydroxylases involved in the formation of threo- and erythro-3-OH-Glu, respectively.6 They also confirmed the sequential chlorination of L-Trp by the oxygen- and FADH2-dependent halogenases KtzQ and KtzR involved in the production of diClPIC.7 Finally, they demonstrated that KtzA-D are involved in the formation of (–)-(1S,2R)-allocoronamic acid instead of the originally proposed D-mecPG.8 Although much progress has been made toward understanding kutznerides biosynthesis, the formation of the other key component of the kutznerides core, O-Me-L-Ser, remains unexplored.
As our group is interested in understanding and engineering unique adenylation (A) domains for future combinatorial biosynthesis,9, 10, 11 we were intrigued by the formation of O-Me-L-Ser by this potential activation and O-methylation of L-Ser by the adenylation domain (A4) of KtzH, which is interrupted by a part of a methyltransferase (M) enzyme (Figure 1c). MbtH-like proteins were recently demonstrated to have a critical role as beneficial and, in some cases, obligatory A domain partners required for the solubility and activity of these A domains.12, 13, 14, 15, 16, 17, 18 Although still controversial, out of the proposed roles for MbtH-like proteins, folding chaperones,18, 19 integral parts of the nonribosomal peptide synthetase complex,16 or allosteric regulators of adenylating enzymes,20 recent structural studies of an enzyme comprised of an MbtH-like protein and an A domain20 point to the latter as a more plausible role for MtbH-like proteins. We were also interested in understanding the possible importance of the MbtH-like protein KtzJ in production of O-Me-L-Ser. In this study, we report our efforts towards delineating O-Me-L-Ser formation in kutznerides biosynthesis.
Materials and methods
Bacterial strains, plasmids, materials and instrumentation
Primers used for PCR were purchased from Integrated DNA Technologies (Coralville, IA, USA). Restriction enzymes, Phusion DNA polymerase, T4 DNA ligase and all other cloning reagents were purchased from New England Biolabs (Ipswich, MA, USA). Chemically competent E. coli TOP10 was purchased from Invitrogen (Carlsbad, CA, USA). The E. coli BL21 (DE3)ybdZ::aac(3)IV strain was generously provided by Professor Michael G. Thomas (University of Wisconsin-Madison, WI, USA). The pET28a and pACYCDuet-1 vectors were purchased from Novagen (Gibbstown, NJ, USA). All chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA) and used without any further purification. [methyl-3H]SAM (S-adenosylmethionine) and [3H]acetyl-CoA were purchased from American Radiolabeled Chemicals (St Louis, MO, USA). [32P]PPi and [3H(G)]L-Ser were purchased from Perkin Elmer (Waltham, MA, USA). DNA sequencing was performed at the University of Michigan DNA sequencing Core.
Preparation of pKtzH(A4MA4T4)-pET28a and pKtzJ-pACYCDuet-1 overexpression constructs
The genes encoding KtzH(A4MA4T4) and KtzJ were PCR-amplified using Kutzneria sp. 744 genomic DNA (Kutzneria sp. 744 used for genomic DNA isolation was a generous gift from Dr Anders Broberg, Swedish University of Agricultural Sciences, Uppsala, Sweden) and Phusion DNA polymerase as described by New England Biolabs. The primers used for the PCR amplification of KtzH(A4MA4T4) (forward (5′-GCCGCCCATATGACCGTGCCGCTGACCG-3′) and reverse (5′-CAGCGGCTCGAGCTACGGCAGCACCTCGGC-3′)) and KtzJ (forward (5′-AAGGAGGAATTCCATGAGCGCCAACCCGTTC-3′) and reverse (5′-CGGTCCAAGCTTTCAGTCGGCCGCCATGGCCTC-3′)) introduced NdeI/XhoI and EcoRI/HindIII restrictions sites (underlined), respectively. The amplified KtzH(A4MA4T4) and KtzJ genes were inserted into the linearized pET28a and pACYCDuet-1 vectors via the corresponding NdeI/XhoI and EcoRI/HindIII restriction sites, respectively, to give constructs pKtzH(A4MA4T4)-pET28a and pKtzJ-pACYCDuet-1 encoding NHis-tagged proteins. All cloning experiments were done in E. coli TOP10 chemically competent cells. Both expression clones were sequenced; these sequences matched perfectly with the annotated ones (accession numbers ABV56588 (KtzH) and ABV56590 (KtzJ)).
Co-overproduction and purification of KtzH(A4MA4T4) and KtzJ proteins
The purified plasmid pKtzH(A4MA4T4)-pET28a was co-transformed with pKtzJ-pACYCDuet-1 into E. coli BL21 (DE3)ybdZ::aac(3)IV competent cells for protein co-expression and purification. One liter of Luria-Bertani medium supplemented with MgCl2 (10 mM final concentration), kanamycin (50 μg ml−1) and chloramphenicol (25 μg ml−1) was inoculated with 5 ml of an overnight culture of a fresh co-transformant and incubated (25 °C, 200 r.p.m.) until the bacterial culture reached an OD600 of ∼0.7. The culture was then cooled to 15 °C before induction with isopropryl-β-thiogalactopyranoside (0.1 ml of a 1 M stock) and shaken for an additional 16 h at 15 °C. Cells were harvested by centrifugation (6000 r.p.m., 10 min, 4 °C) and resuspended in buffer A (25 mM Tris-HCl pH 8.0, 400 mM NaCl and 10% glycerol). The resuspended cells were lysed by sonication (5 min using 10 s ‘on’ alternating with 20 s ‘off’) and the cell debris was removed by centrifugation (16 000 r.p.m., 45 min, 4 °C). Imidazole (2 mM final concentration) was added to the supernatant before incubation with 3 ml of Ni-NTA agarose resin (Qiagen, Gaithersburg, MD, USA) at 4 °C for 2 h with gentle rocking. The resin was loaded onto a column and washed with 10 ml of buffer A containing 5 mM imidazole and then with 10 ml of buffer A containing 20 mM imidazole. KtzH(A4MA4T4) and KtzJ were co-eluted from the column in a stepwise imidazole gradient (one 5 ml fraction of 20 mM, 40 mM and 60 mM, as well as two 5 ml fractions of 200 mM and 500 mM imidazole). Fractions with 200 mM imidazole containing the desired proteins (as determined by SDS-polyacrylamide gel electrophoresis) were combined and dialyzed at 4 °C overnight against a total of 8 l of buffer B (40 mM Tris-HCl pH 8.0, 200 mM NaCl and 10% glycerol). The KtzH(A4MA4T4) and KtzJ co-eluted proteins were concentrated using Amicon Ultra PL-3 concentrators, flash frozen in liquid nitrogen and stored at −80 °C.
It is important to note that only the co-expression and purification of KtzH(A4MA4T4) with its MbtH-like protein partner KtzJ described in this section yielded soluble and active KtzH(A4MA4T4). The following experiments were also attempted to express and purify KtzH(A4MA4T4) alone but yielded insoluble protein. We generated plasmids pKtzH(A4MA4T4)-pET28a, pKtzH(A4MA4T4)-Int-pET19b-pps, pKtzH(A4MA4T4)-pET22b, pKtzH(A4MA4T4)-pGS-21a, pKtzH(A4MA4T4)-pMCSG7 encoding proteins with NHis6, NHis10, CHis6, GST and MOCR tags, respectively. We attempted to express KtzH(A4MA4T4) from these constructs in BL21 (DE3) at various temperature (15, 25 and 37 °C), inducing with various amounts of isopropryl-β-thiogalactopyranoside (100, 250, 500 and 1000 μM) and growing for different periods (2, 5 and 16 h) after induction. We also attempted different lysis methods (sonication and cell disruption using an Avestin Emulsiflex-C3 (Ottawa, ON, Canada)).
Substrate specificity and determination of kinetic parameters for the A domain of KtzH(A4MA4T4) by ATP-[32P]PPi exchange assays
To establish the substrate specificity profile of the A domain of KtzH(A4MA4T4), ATP-[32P]PPi exchange assays were performed for 2 h at room temperature (RT) in reactions (100 μl) containing Tris-HCl (75 mM, pH 7.5), TCEP (5 mM, pH 7.0), MgCl2 (10 mM), ATP (5 mM), Na4P2O7 (1 mM, containing ∼400 000 c.p.m. of [32P]PPi per reaction), amino-acid substrate (5 mM) and KtzH(A4MA4T4) (1 μM) (co-expressed and co-purified with KtzJ) as previously described.14 For the determination of the kinetic parameters (Km and kcat) for L-Ser, duplicate reactions (100 μl) containing various concentrations of L-Ser (0, 0.05, 0.1, 0.25, 0.5, 1, 1.75, 2.5, 5, 10 and 17 mM) were initiated by addition of KtzH(A4MA4T4) (1 μM) (co-expressed and co-purified with KtzJ) and stopped after 15 min.
Characterization of the T domain of KtzH(A4MA4T4) in time-limited assays
To confirm the activity of the T domain of KtzH(A4MA4T4), its conversion from its inactive (apo=non-modified T-domain enzyme) to active (holo=T domain, where the active site serine has been modified by addition of a phosphopantetheine arm from the action of a phosphopantetheinyltransferase (for example, Sfp) and CoA) form was first determined by incorporation of [3H]acetyl into the apo protein by using trichloroacetic acid (TCA) precipitation assays at RT as previously described.21 Briefly, reaction mixtures (25 μl) containing Tris-HCl (75 mM, pH 7.5), MgCl2 (10 mM), TCEP (1 mM, pH 7.0), [3H]acetyl-CoA (100 μM), apo KtzH(A4MA4T4) (25 μM) (co-expressed and co-purified with KtzJ) were initiated by addition of Sfp (1 μM) and terminated after 1, 2, 3, 15 and 60 min before processing and liquid scintillation counting.
To confirm loading of L-Ser onto the holo T domain, the apo to holo conversion (12.5 μl) was first performed for 2 h as described above, but by using CoA instead of [3H]acetyl-CoA. Simultaneously, the activation of L-Ser to L-Ser-AMP was performed in a separate reaction mixture (12.5 μl) containing Tris-HCl (75 mM, pH 7.5), MgCl2 (10 mM), TCEP (1 mM, pH 7.0), ATP (5 mM), apo KtzH(A4MA4T4) (5 μM) (co-expressed and co-purified with KtzJ), L-Ser (200 μM, containing ∼400 000 c.p.m. of [3H(G)]L-Ser per reaction). After 2 h of incubation at RT for each individual reaction mixture, the reaction mixtures (12.5 μl each) containing the holo enzyme and the [3H]L-Ser-AMP were combined. Reactions (25 μl total) were terminated after 5, 10, 15, 30, 90 and 180 min by addition of 10% TCA (100 μl). The protein was pelleted by centrifugation (13 000 r.p.m., RT, 7 min), washed with 10% TCA (100 μl) and resuspended in 88% formic acid (100 μl). The radiolabeled product was counted by liquid scintillation counting.
Characterization of the M domain of KtzH(A4MA4T4) in time-limited assays
To confirm the activity of the M domain of KtzH(A4MA4T4), the T domain of the enzyme was first converted from its apo to its holo form in reactions containing Tris-HCl (75 mM, pH 7.5), MgCl2 (10 mM), TCEP (1 mM, pH 7.0), CoA (100 μM), apo KtzH(A4MA4T4) (25 μM) (co-expressed and co-purified with KtzJ) and Sfp (1 μM). After 2 h of incubation at RT, ATP (5 mM) and L-Ser (100 μM) were added for activation by the A domain of the enzyme and loading of the amino acid onto the holo T domain. After an additional 2 h of incubation at RT, SAM (100 μM, containing ∼170 000 c.p.m. of [methyl-3H]SAM per reaction) was added. Reactions (25 μl total) were terminated after 1, 2, 5, 10, 20, 30, 60 and 90 min by addition of 10% TCA (100 μl) and processed as described in the previous section.
Methylation of L-Ser-AMP before loading onto the T domain was also attempted. Reactions containing Tris-HCl (75 mM, pH 7.5), MgCl2 (10 mM), TCEP (1 mM, pH 7.0), ATP (5 mM), apo KtzH(A4MA4T4) (5 μM) (co-expressed and co-purified with KtzJ) and L-Ser were incubated for 2 h at RT. SAM (100 μM, containing ∼170 000 c.p.m. of [methyl-3H]SAM per reaction) was then added. After an additional 2 h of incubation at RT, the holo T domain was added. Reactions (25 μl total) were terminated after 1, 2, 5, 10, 20, 30, 60 and 90 min by addition of 10% TCA (100 μl) and processed as described in the previous section.
Results and discussion
Requirement of the MbtH-like protein KtzJ for KtzH(A4MA4T4) expression and purification
By gene deletion studies, MbtH-like proteins22 were originally demonstrated to be essential for the production of some secondary metabolites.23, 24 In the following years, the importance of these small (∼8 kDa) proteins in assisting A domain production and activity was reported.12, 13, 14, 15, 16, 17, 18 Although structures of MbtH-like proteins have been determined,25, 26 the insight into the structural basis of the interaction of MtbH-like proteins with adenylating enzymes was gained only very recently through the crystal structure of SlgN1, an enzyme composed of an N-terminal MbtH-like protein and a C-terminal A domain.20 The kutznerides gene cluster also contains an MbtH-like protein, KtzJ, that could have a role in production and activity of the interrupted A domain (KtzH(A4MA4)) involved in O-Me-L-Ser formation. To probe the role of KtzJ in KtzH(A4MA4T4) expression, we first attempted to purify KtzH(A4MA4T4) alone, which resulted in no soluble protein production despite all efforts at varying growth temperature, induction conditions, tags (MOCR, His, GST) and tag locations. Expression of soluble and active recombinant KtzH(A4MA4T4) in E. coli BL21 (DE3) was achieved only when we co-expressed it with the MbtH-like protein KtzJ. The co-expressed KtzH(A4MA4T4) and KtzJ were purified by NiII-NTA affinity chromatography (Figure 2) and used in biochemical assays.
Substrate specificity and kinetic characterization of the interrupted A domain of KtzH(A4MA4T4)
During nonribosomal peptide biosynthesis, A domains dictate the identity of the amino acid/amino acid-like building block to be activated for loading onto the downstream thiolation (T)-domain partner and incorporation of this amino-acid residue into the growing peptide chain. A domains are characterized by ten conserved core signature sequences (a1–a10).27 A domains interrupted by a part of a methyltransferase (M),28, 29, 30 a ketoreductase (KR),31 an oxidase (Ox),32, 33 or a monooxygenase (MOx)32, 33 domain have been found in nature. Most commonly, the interruption of A domains occurs between core signature sequences a8 and a9, as in the case of KtzH(A4MA4) (Figure 1b), although interruptions between a2 and a3 as well as between a4 and a5 have also been reported. In the majority of cases of A domains interrupted by M, only the portion of the methylating enzyme containing the SAM-binding sequence is present between two of the ten core signature sequences of the A domain.
To evaluate the proposed role of KtzH(A4MA4T4) in O-Me-L-Ser formation, we determined the substrate specificity profile of the A domain of this enzyme by monitoring the formation of amino acid-AMP by using the well-established ATP-[32P]PPi exchange assay (Figure 3). We confirmed that L-Ser is the substrate of choice for KtzH(A4MA4T4), as none of the 21 other amino acids that were tested acted as substrates of the enzyme. We also established that adenylation of L-Ser occurred before its methylation, as O-methyl-D,L-Ser was not a substrate of KtzH(A4MA4T4).
We also determined the Michaelis–Menten kinetic parameters (Km and kcat) of the steady-state ATP-[32P]PPi exchange by KtzH(A4MA4T4) co-expressed with KtzJ with L-Ser by varying the concentration L-Ser from 0 to 17 mM while keeping the concentration of the enzyme constant at 1 μM (Figure 4). The Km value of 3.13±0.26 mM observed for L-Ser is characteristic of the high-μM to low-mM Km values generally observed with isolated A domains. Furthermore, with the kcat value of 2.41±0.07 min−1, the resulting catalytic efficiency of adenylation of L-Ser by the interrupted A domain of KtzH(A4MA4T4) (0.77±0.07 mM−1 min−1) is lower than what is generally observed with uninterrupted A domains (for example, kcat/Km for L-Phe adenylation by TioK co-expressed with TioT=49±8 mM−1 min−1;14 L-Tyr adenylation by CloH co-expressed with CloY=2,670 s−1 M−1;16 and L-Tyr adenylation by SimH co-expressed with SimY=6,180 s−1 M−1 16). These data suggest that, although still active, the dual function of interrupted A domains may come at a cost of efficiency of amino-acid activation.
Activity of the T domain of KtzH(A4MA4T4)
After activation by A domains, the amino acid-AMP are typically transferred to a cognate T domain via covalent tethering to the phosphopantetheine (Ppant) arm of the holo enzyme. To determine whether the L-Ser activated by the interrupted A domain of KtzH(A4MA4T4) could be transferred to T4, we initially monitored the apo to [3H]acetyl-S-T4 conversion of KtzH(A4MA4T4) by the TCA precipitation assay (Figure 5a). With the activity of T4 established, we next observed the transfer of [3H]L-Ser-AMP generated by A4MA4 of KtzH(A4MA4T4) onto the T domain via a similar TCA precipitation assay (Figure 5b).
Methyltransferase activity of the partial M domain of KtzH(A4MA4T4)
Because we could not generate O-methyl-Ser-AMP by using the interrupted A domain in the presence of ATP and O-methyl-D,L-Ser, as it is not a substrate for the enzyme (Figure 3), and as we could not readily generate O-methyl-L-Ser-AMP from L-Ser-AMP in the presence of SAM, we opted to investigate the methyltransferase activity of the partial M domain of KtzH(A4MA4T4) by monitoring the conversion of L-Ser-S-KtzH(A4MA4T4) to [3H]O-methyl-L-Ser-S-KtzH(A4MA4T4) by TCA precipitation assays. After formation of L-Ser-AMP (Figures 3 and 4) and its loading onto T4 (Figure 5), we used [methyl-3H]SAM to methylate L-Ser-S-KtzH(A4MA4T4) and form [3H]O-methyl-L-Ser-S-KtzH(A4MA4T4) (Figure 6). The successful methylation of L-Ser on the loaded T4 domain of KtzH(A4MA4T4) observed in these assays completed the reconstitution of O-Me-L-Ser found in kutznerides.
Conversion of L-Ser-S-KtzH(A4MA4T4) to [3H]O-methyl-L-Ser-S-KtzH(A4MA4T4) observed by TCA precipitation assays. Here, after activation of L-Ser to L-Ser-AMP and subsequent loading of the activated amino acid onto the holo T domain [methyl-3H]SAM was added resulting in the methylation by the M domain of this enzyme.
Conclusions
In summary, we have demonstrated the KtzJ-dependent L-Ser activation and O-methylation by KtzH for kutznerides biosynthesis. We have characterized the activities of the interrupted A/methyltransferase (M) and T domains of KtzH(A4MA4T4), thereby reconstituting the production of O-Me-L-Ser. Even though not as efficient as uninterrupted A domains, A4 domain interrupted by the SAM-binding-containing part of a methyltransferase can perform both the conversion of L-Ser into L-Ser-AMP before loading onto T4 as well as methylation of the amino acid after its covalent attachment to the enzyme. Interrupted A domains present an interesting avenue for generating novel building blocks for combinatorial biosynthesis. We are currently studying interrupted A domains from other biosynthetic gene clusters and working toward engineering novel enzymes for future amino-acid derivatives production.
References
Broberg, A., Menkis, A. & Vasiliauskas, R. Kutznerides 1-4, depsipeptides from the actinomycete Kutzneria sp. 744 inhabiting mycorrhizal roots of Picea abies seedlings. J. Nat. Prod. 69, 97–102 (2006).
Pohanka, A., Menkis, A., Levenfors, J. & Broberg, A. Low-abundance kutznerides from Kutzneria sp. 744. J. Nat. Prod. 69, 1776–1781 (2006).
Fujimori, D. G. et al. Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc. Natl Acad. Sci. USA 104, 16498–16503 (2007).
Neumann, C. S. et al. Biosynthesis of piperazic acid via N5-hydroxy-ornithine in Kutzneria spp. 744. Chembiochem 13, 972–976 (2012).
Jiang, W. et al. Biosynthetic chlorination of the piperazate residue in kutzneride biosynthesis by KthP. Biochemistry 50, 6063–6072 (2011).
Strieker, M., Nolan, E. M., Walsh, C. T. & Marahiel, M. A. Stereospecific synthesis of threo- and erythro-beta-hydroxyglutamic acid during kutzneride biosynthesis. J. Am. Chem. Soc. 131, 13523–13530 (2009).
Heemstra, J. R. Jr. & Walsh, C. T. Tandem action of the O2- and FADH2-dependent halogenases KtzQ and KtzR produce 6,7-dichlorotryptophan for kutzneride assembly. J. Am. Chem. Soc. 130, 14024–14025 (2008).
Neumann, C. S. & Walsh, C. T. Biosynthesis of (−)-(1S,2R)-allocoronamic acyl thioester by an Fe(II)-dependent halogenase and a cyclopropane-forming flavoprotein. J. Am. Chem. Soc. 130, 14022–14023 (2008).
McQuade, T. J. et al. A nonradioactive high-throughput assay for screening and characterization of adenylation domains for nonribosomal peptide combinatorial biosynthesis. Anal. Biochem. 386, 244–250 (2009).
Garneau-Tsodikova, S., Dorrestein, P. C., Kelleher, N. L. & Walsh, C. T. Protein assembly line components in prodigiosin biosynthesis: characterization of PigA,G,H,I,J. J. Am. Chem. Soc. 128, 12600–12601 (2006).
Garneau, S., Dorrestein, P. C., Kelleher, N. L. & Walsh, C. T. Characterization of the formation of the pyrrole moiety during clorobiocin and coumermycin A1 biosynthesis. Biochemistry 44, 2770–2780 (2005).
Felnagle, E. A. et al. MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry 49, 8815–8817 (2010).
Zhang, W., Heemstra, J. R. Jr., Walsh, C. T. & Imker, H. J. Activation of the pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry 49, 9946–9947 (2010).
Zolova, O. E. & Garneau-Tsodikova, S. Importance of the MbtH-like protein TioT in production and activation of the thiocoraline adenylation domain of TioK. Med. Chem. Comm. 3, 950–955 (2012).
Zhang, C. et al. In vitro characterization of echinomycin biosynthesis: formation and hydroxylation of L-tryptophanyl-S-enzyme and oxidation of (2S,3S) beta-hydroxytryptophan. PLoS One 8, e56772 (2013).
Boll, B., Taubitz, T. & Heide, L. Role of MbtH-like proteins in the adenylation of tyrosine during aminocoumarin and vancomycin biosynthesis. J. Biol. Chem. 286, 36281–36290 (2011).
Davidsen, J. M., Bartley, D. M. & Townsend, C. A. Non-ribosomal propeptide precursor in nocardicin A biosynthesis predicted from adenylation domain specificity dependent on the MbtH family protein NocI. J. Am. Chem. Soc. 135, 1749–1759 (2013).
McMahon, M. D., Rush, J. S. & Thomas, M. G. Analyses of MbtB, MbtE, and MbtF suggest revisions to the mycobactin biosynthesis pathway in Mycobacterium tuberculosis. J. Bacteriol. 194, 2809–2818 (2012).
Imker, H. J., Krahn, D., Clerc, J., Kaiser, M. & Walsh, C. T. N-acylation during glidobactin biosynthesis by the tridomain nonribosomal peptide synthetase module GlbF. Chem. Biol. 17, 1077–1083 (2010).
Herbst, D. A., Boll, B., Zocher, G., Stehle, T. & Heide, L. Structural basis of the interaction of MbtH-like proteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes. J. Biol. Chem. 288, 1991–2003 (2013).
Mady, A. S. et al. Characterization of TioQ, a type II thioesterase from the thiocoraline biosynthetic cluster. Mol. Biosyst. 7, 1999–2011 (2011).
Baltz, R. H. Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J. Ind. Microbiol. Biotechnol. 38, 1747–1760 (2011).
Wolpert, M., Gust, B., Kammerer, B. & Heide, L. Effects of deletions of mbtH-like genes on clorobiocin biosynthesis in Streptomyces coelicolor. Microbiology 153 (Pt 5), 1413–1423 (2007).
Lautru, S., Oves-Costales, D., Pernodet, J. L. & Challis, G. L. MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145. Microbiology 153 (Pt 5), 1405–1412 (2007).
Drake, E. J. et al. The 1.8 A crystal structure of PA2412, an MbtH-like protein from the pyoverdine cluster of Pseudomonas aeruginosa. J. Biol. Chem. 282, 20425–20434 (2007).
Buchko, G. W., Kim, C. Y., Terwilliger, T. C. & Myler, P. J. Solution structure of Rv2377c-founding member of the MbtH-like protein family. Tuberculosis 90, 245–251 (2010).
Gulick, A. M. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS. Chem. Biol. 4, 811–827 (2009).
Zolova, O. E., Mady, A. S. & Garneau-Tsodikova, S. Recent developments in bisintercalator natural products. Biopolymers 93, 777–790 (2010).
Patel, H. M. & Walsh, C. T. In vitro reconstitution of the Pseudomonas aeruginosa nonribosomal peptide synthesis of pyochelin: characterization of backbone tailoring thiazoline reductase and N-methyltransferase activities. Biochemistry 40, 9023–9031 (2001).
Sandmann, A., Sasse, F. & Muller, R. Identification and analysis of the core biosynthetic machinery of tubulysin, a potent cytotoxin with potential anticancer activity. Chem. Biol. 11, 1071–1079 (2004).
Magarvey, N. A., Ehling-Schulz, M. & Walsh, C. T. Characterization of the cereulide NRPS alpha-hydroxy acid specifying modules: activation of alpha-keto acids and chiral reduction on the assembly line. J. Am. Chem. Soc. 128, 10698–10699 (2006).
Silakowski, B. et al. New lessons for combinatorial biosynthesis from myxobacteria. The myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 274, 37391–37399 (1999).
Weinig, S., Hecht, H. J., Mahmud, T. & Muller, R. Melithiazol biosynthesis: further insights into myxobacterial PKS/NRPS systems and evidence for a new subclass of methyl transferases. Chem. Biol. 10, 939–952 (2003).
Acknowledgements
This work was supported by a NSF CAREER Award (grant no. MCB-1149427) (to SG-T) as well as by startup funds from the University of Kentucky (to SG-T). We thank Finn P. Maloney and Dr Wenjing Chen for cloning of the KtzH and KtzJ constructs used in this study. We thank Dr Oleg V. Tsodikov for critical reading of the manuscript and insightful comments. We thank Dr Anders Broberg (Swedish University of Agricultural Sciences, Uppsala, Sweden) for the gift of the Kutzneria sp. 744 bacterial strain used to isolate genomic DNA. We thank Dr Michael G. Thomas (University of Wisconsin-Madison, WI, USA) for the gift of the BL21 (DE3)ybdZ::aac(3)IV bacterial strain. SG-T thanks Professor Christopher T. Walsh, not only an amazing mentor but also a continuous source of inspiration and support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interest.
Rights and permissions
About this article
Cite this article
Zolova, O., Garneau-Tsodikova, S. KtzJ-dependent serine activation and O-methylation by KtzH for kutznerides biosynthesis. J Antibiot 67, 59–64 (2014). https://doi.org/10.1038/ja.2013.98
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.98
Keywords
This article is cited by
-
Antifungal peptides produced by actinomycetes and their biological activities against plant diseases
The Journal of Antibiotics (2020)
-
Refining and expanding nonribosomal peptide synthetase function and mechanism
Journal of Industrial Microbiology and Biotechnology (2019)