Abstract
An actinomycete strain, S3-1T, was isolated from marine sponge sample collected from the Gulf of Thailand. The strain is aerobic, Gram-positive and produced single spores at the tip of the substrate mycelium. Strain S3-1T contained meso-diaminopimelic acid in the peptidoglycan, whole-cell sugars were arabinose, galactose, glucose, rhamnose, ribose and xylose. The polar lipid profile of strain S3-1T consisted of phosphatidylethanolamine, phosphatidylmethylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, phosphoglycolipid and unknown polar lipids. Morphological and chemotaxonomic characteristics of the strain were identified as a member of the genus Micromonospora. Phylogenetic analysis based on 16S rRNA gene sequence analysis of the strain showed similarity to Micromonospora nigra DSM 43818T (98.8%), Micromonospora yangpuensis FXJ6.011T (98.7%) and Micromonospora narathiwatensis BTG4-1T (98.6%). The DNA G+C content was 72.7 mol%. The phenotypic characteristics and DNA–DNA relatedness values supported that the classification of this strain as a novel species in the genus Micromonospora, for which the name Micromonospora spongicola sp. nov. (type strain S3-1T =BCC 45595T=NBRC 108779T) is proposed.
Similar content being viewed by others
Introduction
The genus Micromonospora was proposed by Orskov,1 which belongs to the family Micromonosporaceae.2 The genus can be distinguished from the other member of its family based on the morphological,3 chemotaxonomic and genotypic characteristics.4 Micromonosporae form single non-motile spores on the substrate mycelium, which do not fragment and have absence of aerial mycelium. The genus Micromonospora is probably the second most prolific producer of antibiotics after Streptomyces5 and the first antimicrobial activity by a member of this genus was isolated from Micromonospora echinospora.6 The genus Micromonospora has gathered attention due to the secondary metabolites production with either structural diversity or significant biological activities; for example, Lomaiviticins,7 Lupinacidin8 and Maklamicin,9 in addition, novel compounds exhibiting antitumor and/or antibacterial activities have been also isolated from marine Micromonospora, including Diazepinomicin10 and anthracyclinones.11 Members of Micromonospora are widely distributed in terrestrial, aquatic and marine environments,3, 12, 13 but recently members of this genus have also been recovered from marine sponge.14 In this study, we described the isolation and taxonomic characterization of a novel strain S3-1T, which was isolated from marine sponge in the Gulf of Thailand.
Materials and methods
Strain S3-1T was isolated from marine sponge collected from the Gulf of Thailand by scuba diving at a depth of 5 m in the Sichang Island in Chonburi province. The strain was isolated using modified starch-casein nitrate sea water agar.15 The isolation plate was incubated at 30 °C for 21 days. Actinomycete isolate was purified on ISP 2 medium. Strain S3-1T was grown on soil extract agar medium for 21 days at 30 °C and observed by light microscopy and scanning electron microscopy (model LEO/1455VP). Samples for scanning electron microscopy were prepared as described previously.16
Cultural characteristics were determined using 14 day-cultures grown at 30 °C on several standard agar media. The ISCC–NBS Color Charts standard sample no. 2106 was used for determining color designations.17 Carbon utilization medium (ISP 9)18 supplemented with a final concentration of 1% of the carbon source and 0.05% casamino acids was used to determine the carbon utilization of the strain. The decomposition of various compounds and acid production from carbon sources were examined using the basal medium recommended by Gordon et al.19 To determine the tolerance of NaCl, pH and the effect of temperature for growth were determined on ISP 2 medium. Gelatin liquefaction, peptonization of milk, nitrate reduction and starch hydrolysis were determined through cultivation on various media as described by Arai20 and Williams and Cross.21
Freeze-dried cells used for chemotaxonomic analyses were obtained from cultures grown in ISP 2 broth on rotary shaker (200 r.p.m.) at 30 °C for 7 days. The isomer of diaminopimelic acid in the cell wall was determined by the method of Staneck and Roberts.22 The acyl group of muramic acid in the peptidoglycan was determined by the method of Uchida and Aida.23 The whole-cell hydrolysate sugars were analyzed by the cellulose TLC method of Komagata and Suzuki.24 Polar lipids were extracted and identified by the method of Minnikin et al.25 Fatty acid methyl ester analysis was performed by GLC according to the instructions of the Microbial Identification System (MIDI, version 6.0) (Sasser,26 and Kämpfer and Kroppenstedt27) with the ACTIN1 MIDI database. Isoprenoid quinones were extracted by the method of Collins et al.28 and were examined by reverse phase LC-MS employing UV detection and electrospray MS. The LC solvent system was methanol and 2-propanol (2:1, v/v).
Genomic DNA was isolated from cells grown in ISP 2 broth according to the method of Tamaoka.29 The G+C content (mol%) of the genomic DNA was determined using the HPLC method of Tamaoka and Komagata.30 An equimolar mixture of nucleotides for analysis of the DNA base composition (Yamasa Shoyu) was used as the quantitative standard. DNA–DNA hybridization was conducted in microdilution-well plates, as reported by Ezaki et al.31 DNA–DNA relatedness (%) was determined by the colorimetric method.32 PCR-mediated amplification of the 16S rRNA gene33 and sequencing of the PCR products (Macrogen, Seoul, Korea) using universal primers.34 The 16S rRNA gene sequence was aligned with selected sequences obtained from the GenBank/EMBL/DDBJ databases by using the CLUSTAL W programme version 1.81.35 The alignment was manually verified and adjusted before the construction of a phylogenetic tree. The phylogenetic tree was constructed by using the neighbor-joining36 with genetic distances computed by using Kimura’s 2-parameter model,37 maximum parsimony38 and maximum-likelihood39 methods in the MEGA 5 software.40 The confidence values of branches of the phylogenetic tree were determined using bootstrap analyses41 based on 1000 resamplings. The values for sequence similarity among all recognized Micromonospora species were first determined using the EzTaxon-e database.42 16S rRNA gene sequence similarities among closely related species were calculated manually after pairwise alignments obtained using the CLUSTAL_X program.43
Results and discussion
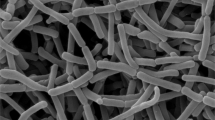
Strain S3-1T formed well-developed and branched substrate mycelia but lacked aerial mycelia. Oval spores were borne singly from the substrate mycelium and the spore surface appeared smooth (Figure 1). The growth of the strain was good on ISP 2 medium and the color of colony of strain S3-1T was light to strong orange but the colony on czapek’s sucrose agar was colorless. Light to strong orange soluble pigment was produced in various media but not produced in czapek’s sucrose agar (Table 1). The temperature and pH range for growth were 20–40 °C and pH 7–12, with optimum growth at 30 °C and pH 8, respectively. The maximum NaCl concentration for growth is 4% (w/v). Other physiological characteristics are given in Table 2 and in the species description.
Strain S3-1T contained meso-diaminopimelic acid as the diagnostic peptidoglycan diamino acid. The acyl type of the cell wall peptidoglycan was determined to be the glycolyl type. Whole-cell hydrolysates contained arabinose, galactose, glucose, rhamnose (trace), ribose and xylose that were detected as whole-cell sugars in this strain. Analysis of polar lipids revealed that the strain contained phosphatidylethanolamine, phosphatidylmethylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, phosphoglycolipid and unknown polar lipids. The major fatty acids (>5%) detected in strain S3-1T was iso-C16:0 (38.1%), anteiso-C17:0 (19.2%), C18:1 ω9c (9.5%), iso-C17:0 (7.2%) and C18:0 (7.2%) (Table 3). The predominant menaquinones were MK-10(H4) (65.2%) and MK-10(H6) (34.8%). The G+C content was 72.7 mol%.
The almost-complete 16S rRNA gene sequence (1457 nucleotides) was obtained for strain S3-1T and compared with those deposited in the public databases. The results indicated that this strain belongs to the genus Micromonospora and showed the highest similarity value was observed with Micromonospora nigra DSM 43818T (98.8%) followed by Micromonospora yangpuensis FXJ6.011T (98.7%) and Micromonospora narathiwatensis BTG4-1T (98.6%). The phylogenetic tree constructed with 16S rRNA gene sequences data of all members of the genus Micromonospora also indicated that strain S3-1T formed a cluster with Micromonospora nigra DSM 43818T and Micromonospora yangpuensis FXJ6.011T using the neighbor-joining method (Figure 2). DNA–DNA hybridization studies performed between strain S3-1T and the related strains Micromonospora nigra NBRC 16103T, Micromonospora yangpuensis NBRC 107727T and Micromonospora narathiwatensis BTG4-1T were 25.0±0.7%, 19.4±1.5% and 14.8±0.9%, respectively.
Phylogenetic dendrogram obtained by neighbour-joining tree based on almost-complete 16S rRNA gene sequences showing the position of strain S3-1T among related species in the Micromonospora species. Catellatospora citrea DSM 44097T was used as an outgroup. Asterisks (*) indicating the branches of the tree that were also found using the neighbour-joining, maximum-parsimony and maximum-likelihood methods; hashes (#) indicating the branches of the tree that were found in the neighbour-joining and maximum-parsimony methods; and crosses (x) indicating the branches of the tree that were found in the neighbor-joining and maximum-likelihood methods. The numbers on the branches indicate the percentage bootstrap values of 1000 replicates; only values >50% are indicated. Bar, 0.005 substitutions per nucleotide position.
The morphological and chemotaxonomic analyzes indicated that strain S3-1T was identified as a member of the genus Micromonospora. However, the 16S rRNA gene sequence similarity values are low (98.6–98.8%) and DNA–DNA relatedness value between strain S3-1T and the closed strains were below 25.0%, less than the value of 70% cutoff point recommended for the assignment of bacterial strains to the same genomic species.44 In addition, the strain S3-1T is distinguished from related strains by differences in the utilization of inositol, D-fructose, sorbose, D-xylose, salicin, xylitol, L-arabitol, L-rhamnose, glycerol, sorbitol and lactose; acid production from D-cellobiose, D-fructose, D-galactose, D-melibiose, D-raffinose, D-xylose, L-arabinose, D-sucrose, lactose, sorbose and salicin. The 10-methyl-C18:0, C14:0 and iso-C14:0 were not detected in strain S3-1T with differentiating properties from these related species. These results supported that strain S3-1T represents a novel species in the genus Micromonospora, for which the name Micromonospora spongicola sp. nov. is proposed.
Description of Micromonospora spongicola sp. nov.
Micromonospora spongicola (spon.gíco.la. L. n. spongos-i sponge; L. suff. -cola (from L. n. incola) inhabitant; N. L. n. (nominative in apposition) spongicola inhabitant of a sponge.
Aerobic, Gram-positive, which form oval spores on substrate mycelium. Aerial mycelium is absent. The color of the substrate mycelium on ISP 2, 3 and glucose-asparagine medium agar is strong orange. Moderate orange soluble pigment is produced on ISP 2, 3, 7 and nutrient medium agar. The maximum temperature for growth is 40 °C. The pH range for growth is 7.0–12.0. The maximum NaCl tolerance is 4% (w/v). L-arabitol, D-fructose, glycerol, inositol, lactose, D-xylose, L-rhamnose, salicin, sorbitol and sorbose are utilized as sole carbon sources but not D-mannitol, D-raffinose, D-ribose, D-mannose and xylitol. In addition, strain S3-1T produced acid from D-cellobiose, D-fructose, D-galactose, lactose, D-melibiose, salicin, sorbose and D-xylose. Starch hydrolysis, gelatin liquefaction and milk peptonization are positive. Nitrate is not reduced. The diagnostic diamino acid of the peptidoglycan is meso-diaminopimelic acid. Whole-cell sugars are arabinose, galactose, glucose, rhamnose, ribose and xylose. Major polar lipids are phosphatidylethanolamine, phosphatidylmethylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, phosphoglycolipid and unknown polar lipids. The fatty acid pattern consists of iso-C16:0, anteiso-C17:0, C18:1 ω9c, iso-C17:0 and C18:0. The predominant menaquinones are MK-10(H4) and MK-10(H6). The DNA G+C content of the type strain is 72.7 mol%. The type strain is S3-1T =BCC 45595T =NBRC 108779T, isolated from the marine sponge collected from the Gulf of Thailand.
Accession code
The DDBJ accession number for the 16S rRNA gene sequence of strain S3-1T is AB645957.
References
Orskov, J. Investigations into the Morphology of the Ray Fungi, Levin and Munksgaard: Copenhagen, (1923).
Stackebrandt, E., Rainey, F. A. & Ward-Rainey, N. L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47, 479–491 (1997).
Kawamoto, I. Genus Micromonospora Orskov 1923, 147AL In. Bergey’s Manual of Systematic Bacteriology edsWilliams S. T., Sharpe M. E., Holt J. G.), vol. 4, pp 2442–2450 Williams & Wilkins: Baltimore, (1989).
Koch, C., Kroppenstedt, R. M., Rainey, F. A. & Stackebrandt, E. 16S ribosomal DNA analysis of the genera Micromonospora, Actinoplanes, Catellatospora, Catenuloplanes, Couchioplanes, Dactylosporangium, and Pilimelia and emendation of the family Micromonosporaceae. Int. J. Syst. Bacteriol. 46, 765–768 (1996).
Wagman, G. H. & Weinstein, M. J. Antibiotic from Micromonospora. Annu. Rev. Microbiol. 34, 537–557 (1980).
Kasai, H., Tamura, T. & Harayama, S. Intrageneric relationships among Micromonospora species deduced from gyrB-based phylogeny and DNA relatedness. Int. J. Syst. Evol. Microbiol. 50, 127–134 (2000).
He, H. et al. Lomaiviticins A and B, potent antitumor antibiotics from Micromonospora lomaiitiensis. Am. Chem. Soc. 123, 5362–5363 (2001).
Igarashi, Y. et al. Lupinacidin C, an inhibitor of tumor cell invasion from Micromonospora lupine. J. Nat. Prod. 74, 862–865 (2011).
Igarashi, Y. et al. Maklamicin, an antibacterial Polyketide from an endophytic Micromonospora sp. J. Nat. Prod. 74, 670–674 (2011).
Charan, R. D. et al. Diazepinomicin, a new antimicrobial alkaloid froma marine Micromonospora sp. J. Nat. Prod. 67, 1431–1433 (2004).
Sousa, T. S. et al. Anthracyclinones from Micromonospora sp. J. Nat. Prod. 75, 489–493 (2012).
Zhao, H., Kassama, Y., Young, M., Kell, D. B. & Goodacre, R. Differentiation of Micromonospora isolates from a coastal sediment in Waleson the basis of Fourier transform infrared spectroscopy 16S rRNA sequence analysis, and the amplified fragment length polymorphism technique. Appl. Environ. Microbiol. 70, 6619–6627 (2004).
Maldonado, L. A. et al. Diversity of cultivable actinobacteria in geographically wide spread marine sediments. Antonie Van Leeuwenhoek 87, 11–18 (2005).
Limin, Z., Lijun, X., Jisheng, R. & Ying, H. Micromonospora yangpuensis sp. nov., isolated from a sponge in South China Sea. Int. J. Syst. Bacteriol. 62, 272–278 (2012).
Supong, K. et al. Micromonospora sediminicola sp. nov., isolated from a marine sediment of the Andaman Sea of Thailand. Int. J. Syst. Bacteriol. 63, 570–575 (2013).
Itoh, T., Kudo, T., Parenti, F. & Seino, A. Amended description of the genus Kineosporia, based on chemotaxonomic and morphological studies. Int. J. Syst. Bacteriol. 39, 168–173 (1989).
Kelly, K. L. Inter-Society Color Council–National Bureau of Standard Color Name Charts Illustrated with Centroid Colors, US Government Printing Office: Washington, DC, (1964).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Gordon, R. E., Barnett, D. A., Handerhan, J. E. & Pang, C. H. N. Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int. J. Syst. Bacteriol. 24, 54–63 (1974).
Arai, T. Culture Media for Actinomycetes., Tokyo: : The Society for Actinomycetes, Japan, ((1975).
Williams, S. T. & Cross, T. Actinomycetes In Methods in Microbiology (edBooth C.), vol. 4, pp 295–334 Academic Press: London, (1971).
Staneck, J. L. & Roberts, G. D. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231 (1974).
Uchida, K. & Aida, K. An improved method for the glycolate test for simple identification of the acyl type of bacterial cell walls. J. Gen. Appl. Microbiol. 30, 131–134 (1984).
Komagata, K. & Suzuki, K. I. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 19, 161–207 (1987).
Minnikin, D. E. et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241 (1984).
Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids (MIDI Technical Note 101), MIDI: Newark, DE, ((1990).
Kämpfer, P. & Kroppenstedt, R. M. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can. J. Microbiol. 42, 989–1005 (1996).
Collins, M. D., Pirouz, T., Goodfellow, M. & Minnikin, D. E. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230 (1977).
Tamaoka, J. Determination of DNA Base Composition In Chemical Methods in Prokaryotic Systematics edsGoodfellow M., O’Donnell A. G., pp 463–470 John Wiley & Sons: Chichester, (1994).
Tamaoka, J. & Komagata, K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128 (1984).
Ezaki, T., Hashimoto, Y. & Yabuuchi, E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in micro-dilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229 (1989).
Verlander, C. P. Detection of horseradish peroxidase by colorimetry In Nonisotopic DNA Probe Techniques edKricka L. J., pp 185–201 Academic Press: New York, (1992).
Suriyachadkun, C. et al. Planotetraspora thailandica sp. nov., isolated from soil in Thailand. Int. J. Syst. Evol. Microbiol. 59, 992–997 (2009).
Lane, D. J. 16S/23S rRNA sequencing In Nucleic Acid Techniques in Bacterial Systematics edsStackebrandt E., Goodfellow M., pp 115–148 John Wiley & Sons: Chichester, (1991).
Thompson, J. D., Higgins, D. G. & Gobson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Fitch, W. M. Toward defining the course of evolution: minimum change for a species tree topology. Sys. Zoo. 20, 406–416 (1971).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Kim, O. S. et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Wayne, L. G. et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463464 (1987).
Acknowledgements
This study was financially supported by the research fund from the Thailand Graduate Institute of Science and Technology (TGIST-01-52-003), National Science and Technology Development Agency (NSTDA) to S. K. is gratefully acknowledged. This study was supported in part by Department of Biology and Actinobacterial Research Unit, Faculty of Science, King Mongkut’s Institute of Technology, Ladkrabang, Bangkok, Thailand.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Supong, K., Suriyachadkun, C., Pittayakhajonwut, P. et al. Micromonospora spongicola sp. nov., an actinomycete isolated from a marine sponge in the Gulf of Thailand. J Antibiot 66, 505–509 (2013). https://doi.org/10.1038/ja.2013.35
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.35
Keywords
This article is cited by
-
Streptomyces telluris sp. nov., a promising terrestrial actinobacterium with antioxidative potentials
Archives of Microbiology (2023)
-
Uncovering the biodiversity and biosynthetic potentials of rare actinomycetes
Future Journal of Pharmaceutical Sciences (2022)
-
Micromonospora azadirachtae sp. nov., isolated from roots of Azadirachta indica A. Juss. var. siamensis Valeton
Antonie van Leeuwenhoek (2019)
-
Recent progress on the development of antibiotics from the genus Micromonospora
Biotechnology and Bioprocess Engineering (2016)